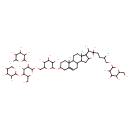| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 03:03:00 UTC |
|---|
| Update Date | 2016-11-09 01:19:22 UTC |
|---|
| Accession Number | CHEM031965 |
|---|
| Identification |
|---|
| Common Name | Trigofoenoside G |
|---|
| Class | Small Molecule |
|---|
| Description | Trigofoenoside G is found in fenugreek. Trigofoenoside G is isolated from fenugreek seeds. |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
| Chemical Structure | |
|---|
| Synonyms | Not Available |
|---|
| Chemical Formula | C56H92O27 |
|---|
| Average Molecular Mass | 1197.314 g/mol |
|---|
| Monoisotopic Mass | 1196.583 g/mol |
|---|
| CAS Registry Number | 94714-57-5 |
|---|
| IUPAC Name | 2-({[4-hydroxy-6-(hydroxymethyl)-3-[(3,4,5-trihydroxy-6-methyloxan-2-yl)oxy]-5-[(3,4,5-trihydroxyoxan-2-yl)oxy]oxan-2-yl]oxy}methyl)-6-{[6-hydroxy-7,9,13-trimethyl-6-(3-methyl-4-{[3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy}butyl)-5-oxapentacyclo[10.8.0.0²,⁹.0⁴,⁸.0¹³,¹⁸]icos-18-en-16-yl]oxy}oxane-3,4,5-triol |
|---|
| Traditional Name | 2-({[4-hydroxy-6-(hydroxymethyl)-3-[(3,4,5-trihydroxy-6-methyloxan-2-yl)oxy]-5-[(3,4,5-trihydroxyoxan-2-yl)oxy]oxan-2-yl]oxy}methyl)-6-{[6-hydroxy-7,9,13-trimethyl-6-(3-methyl-4-{[3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy}butyl)-5-oxapentacyclo[10.8.0.0²,⁹.0⁴,⁸.0¹³,¹⁸]icos-18-en-16-yl]oxy}oxane-3,4,5-triol |
|---|
| SMILES | CC(CCC1(O)OC2CC3C4CC=C5CC(CCC5(C)C4CCC3(C)C2C1C)OC1OC(COC2OC(CO)C(OC3OCC(O)C(O)C3O)C(O)C2OC2OC(C)C(O)C(O)C2O)C(O)C(O)C1O)COC1OC(CO)C(O)C(O)C1O |
|---|
| InChI Identifier | InChI=1S/C56H92O27/c1-21(18-73-49-43(68)40(65)37(62)31(16-57)78-49)8-13-56(72)22(2)34-30(83-56)15-28-26-7-6-24-14-25(9-11-54(24,4)27(26)10-12-55(28,34)5)77-52-45(70)41(66)38(63)33(80-52)20-75-53-48(82-51-44(69)39(64)35(60)23(3)76-51)46(71)47(32(17-58)79-53)81-50-42(67)36(61)29(59)19-74-50/h6,21-23,25-53,57-72H,7-20H2,1-5H3 |
|---|
| InChI Key | BTKYWGCMCQREJD-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as steroidal saponins. These are saponins in which the aglycone moiety is a steroid. The steroidal aglycone is usually a spirostane, furostane, spirosolane, solanidane, or curcubitacin derivative. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Steroids and steroid derivatives |
|---|
| Sub Class | Steroidal glycosides |
|---|
| Direct Parent | Steroidal saponins |
|---|
| Alternative Parents | |
|---|
| Substituents | - Steroidal saponin
- Diterpene glycoside
- Furostane-skeleton
- Oligosaccharide
- 22-hydroxysteroid
- Diterpenoid
- Hydroxysteroid
- Delta-5-steroid
- Terpene glycoside
- Fatty acyl glycoside
- Alkyl glycoside
- Glycosyl compound
- O-glycosyl compound
- Fatty acyl
- Oxane
- Tetrahydrofuran
- Secondary alcohol
- Hemiacetal
- Organoheterocyclic compound
- Oxacycle
- Polyol
- Acetal
- Alcohol
- Hydrocarbon derivative
- Organic oxygen compound
- Primary alcohol
- Organooxygen compound
- Aliphatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aliphatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0032-9600150002-b41cb5342b53b90cb9ab | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00nb-7420390225-175759ce79324a7e582e | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00mk-9622580016-dd7b5c0359dcbfd03d92 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-01r2-4900120001-c26ff6fada43239128cd | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-01t9-2900020000-8a8e82aa61555f37a2e6 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-01ox-5900040001-d4ea43a1df0a2594ed6e | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-002b-2900010000-2ea293b25956b40fc9fb | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-014j-4902030012-cdfa7511378afb569443 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-014m-9820112120-1550fa4b8f70a1f724ad | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-1900000000-dba1a5ae6030a124cbe6 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4m-9700000000-85672a05e78e3938bb62 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-9200001002-07dba241e27b4e2c60ed | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0038616 |
|---|
| FooDB ID | FDB018010 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | Not Available |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 131752408 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|