| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 03:01:00 UTC |
|---|
| Update Date | 2016-11-09 01:19:22 UTC |
|---|
| Accession Number | CHEM031922 |
|---|
| Identification |
|---|
| Common Name | 3'-N-Acetyl-4'-O-(10,12-octadecadienoyl)fusarochromanone |
|---|
| Class | Small Molecule |
|---|
| Description | 3'-N-Acetyl-4'-O-(10,12-octadecadienoyl)fusarochromanone is produced by Fusarium equiseti. |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
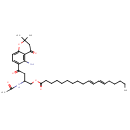
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| N-[4-(5-Amino-2,2-dimethyl-4-oxo-3,4-dihydro-2H-1-benzopyran-6-yl)-1-[(10E,12E)-octadeca-10,12-dienoyloxy]-4-oxobutan-2-yl]ethanimidate | HMDB |
|
|---|
| Chemical Formula | C35H52N2O6 |
|---|
| Average Molecular Mass | 596.797 g/mol |
|---|
| Monoisotopic Mass | 596.383 g/mol |
|---|
| CAS Registry Number | 136536-84-0 |
|---|
| IUPAC Name | 4-(5-amino-2,2-dimethyl-4-oxo-3,4-dihydro-2H-1-benzopyran-6-yl)-2-acetamido-4-oxobutyl (10E,12E)-octadeca-10,12-dienoate |
|---|
| Traditional Name | 4-(5-amino-2,2-dimethyl-4-oxo-3H-1-benzopyran-6-yl)-2-acetamido-4-oxobutyl (10E,12E)-octadeca-10,12-dienoate |
|---|
| SMILES | CCCCC\C=C\C=C\CCCCCCCCC(=O)OCC(CC(=O)C1=CC=C2OC(C)(C)CC(=O)C2=C1N)NC(C)=O |
|---|
| InChI Identifier | InChI=1S/C35H52N2O6/c1-5-6-7-8-9-10-11-12-13-14-15-16-17-18-19-20-32(41)42-25-27(37-26(2)38)23-29(39)28-21-22-31-33(34(28)36)30(40)24-35(3,4)43-31/h9-12,21-22,27H,5-8,13-20,23-25,36H2,1-4H3,(H,37,38)/b10-9+,12-11+ |
|---|
| InChI Key | MCIFNOLGPRTLRK-HULFFUFUSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as lineolic acids and derivatives. These are derivatives of lineolic acid. Lineolic acid is a polyunsaturated omega-6 18 carbon long fatty acid, with two CC double bonds at the 9- and 12-positions. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Fatty Acyls |
|---|
| Sub Class | Lineolic acids and derivatives |
|---|
| Direct Parent | Lineolic acids and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Octadecanoid
- 2,2-dimethyl-1-benzopyran
- Butyrophenone
- Chromone
- Chromane
- Benzopyran
- 1-benzopyran
- Aryl alkyl ketone
- Aryl ketone
- Alkyl aryl ether
- Fatty acid ester
- Benzenoid
- Acetamide
- Vinylogous amide
- Amino acid or derivatives
- Carboxamide group
- Carboxylic acid ester
- Ketone
- Secondary carboxylic acid amide
- Organoheterocyclic compound
- Oxacycle
- Ether
- Carboxylic acid derivative
- Monocarboxylic acid or derivatives
- Hydrocarbon derivative
- Organic nitrogen compound
- Amine
- Carbonyl group
- Organic oxide
- Organopnictogen compound
- Organonitrogen compound
- Organooxygen compound
- Primary amine
- Organic oxygen compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-014r-3292110000-fafdf52fc7763e292e41 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0hit-0031090000-d8a3cf98a678e60bf000 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-014i-0092030000-3767db8f4ff1f058b6c5 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-07vi-5290320000-e71de566873c581de89f | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-01r2-0082090000-7ebd8c12b2e32e6a1222 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-03fr-2092030000-805cba0e186b91002e7f | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0bvl-7090000000-c15adf691d22c206653f | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014j-0039060000-157e616355efa26f1385 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-014i-1197010000-fcada93ef0af6f6ec612 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-014i-2596000000-30e4f56dae0b7d5a2948 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-1032090000-52abfbf33f461c831b9c | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-05vk-3092010000-53420fc048ce8421b20d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-02hc-4090000000-cbc1909dfc8a25dac01b | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0038566 |
|---|
| FooDB ID | FDB017953 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | C00055094 |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 35014604 |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 102145835 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|