| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 02:44:27 UTC |
|---|
| Update Date | 2016-11-09 01:19:17 UTC |
|---|
| Accession Number | CHEM031555 |
|---|
| Identification |
|---|
| Common Name | Absintholide |
|---|
| Class | Small Molecule |
|---|
| Description | Constituent of Artemisia absinthium (wormwood). Absintholide is found in alcoholic beverages and herbs and spices. |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
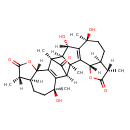
| Chemical Structure | |
|---|
| Synonyms | Not Available |
|---|
| Chemical Formula | C30H38O8 |
|---|
| Average Molecular Mass | 526.618 g/mol |
|---|
| Monoisotopic Mass | 526.257 g/mol |
|---|
| CAS Registry Number | 91997-90-9 |
|---|
| IUPAC Name | (1S,2R,4S,7S,8S,11S,13S,14S,15R,17S,20S,21S,24S)-11,13,24-trihydroxy-2,7,11,15,20,24-hexamethyl-5,18-dioxaheptacyclo[13.10.1.0²,¹⁴.0³,¹².0⁴,⁸.0¹⁶,²⁵.0¹⁷,²¹]hexacosa-3(12),16(25)-diene-6,19,26-trione |
|---|
| Traditional Name | (1S,2R,4S,7S,8S,11S,13S,14S,15R,17S,20S,21S,24S)-11,13,24-trihydroxy-2,7,11,15,20,24-hexamethyl-5,18-dioxaheptacyclo[13.10.1.0²,¹⁴.0³,¹².0⁴,⁸.0¹⁶,²⁵.0¹⁷,²¹]hexacosa-3(12),16(25)-diene-6,19,26-trione |
|---|
| SMILES | [H][C@@]12C(=O)[C@@](C)(C3=C1[C@@](C)(O)CC[C@H]1[C@H](C)C(=O)O[C@H]31)[C@@]1([H])[C@H](O)C3=C([C@H]4OC(=O)[C@@H](C)[C@@H]4CC[C@]3(C)O)[C@@]21C |
|---|
| InChI Identifier | InChI=1S/C30H38O8/c1-11-13-7-9-27(3,35)15-17(21(13)37-25(11)33)30(6)23-20(31)16-18(29(23,5)19(15)24(30)32)22-14(8-10-28(16,4)36)12(2)26(34)38-22/h11-14,19-23,31,35-36H,7-10H2,1-6H3/t11-,12-,13-,14-,19-,20+,21-,22-,23-,27-,28-,29-,30-/m0/s1 |
|---|
| InChI Key | ANVQPXYQHSOZNE-UMDUJLOZSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as sesterterpenoids. These are terpenes composed of five consecutive isoprene units. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Sesterterpenoids |
|---|
| Direct Parent | Sesterterpenoids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Sesterterpenoid
- Prostaglandin skeleton
- Eicosanoid
- Dicarboxylic acid or derivatives
- Gamma butyrolactone
- Fatty acyl
- Tertiary alcohol
- Tetrahydrofuran
- Secondary alcohol
- Lactone
- Ketone
- Carboxylic acid ester
- Organoheterocyclic compound
- Oxacycle
- Polyol
- Carboxylic acid derivative
- Organooxygen compound
- Hydrocarbon derivative
- Organic oxide
- Organic oxygen compound
- Carbonyl group
- Alcohol
- Aliphatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aliphatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-03e9-1048950000-a4131d230bed43d41578 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-0a4i-0001139000-45de5715331a7227886e | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0a4l-0000970000-1fff10de6af71714141b | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-052u-0000920000-e05bc31174207213b909 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000i-0133900000-93a9155fb1764c62b86a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0059-0000690000-d5bf18189ee1a12e5c60 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a59-0000970000-be3c89e11933dcdb98d5 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-002r-0001900000-9edde8842146afffb4ef | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-004i-0000090000-b779f3c387e69eefba3e | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0bu3-0010950000-d4186b51d4ed467afa99 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-014i-0193400000-a480cfbdbd768e08c983 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-0000190000-165cd87e93697615d835 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0059-0000790000-116ad23722c572e5c2b7 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-056s-1010920000-b48723bf689996921892 | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0038171 |
|---|
| FooDB ID | FDB017402 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | C00020967 |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | Not Available |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 101087895 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|