| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 02:39:29 UTC |
|---|
| Update Date | 2016-11-09 01:19:16 UTC |
|---|
| Accession Number | CHEM031444 |
|---|
| Identification |
|---|
| Common Name | Eriojaposide B |
|---|
| Class | Small Molecule |
|---|
| Description | Eriojaposide B is found in fruits. Eriojaposide B is a constituent of Eriobotrya japonica (loquat). |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
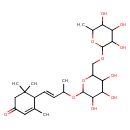
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (6R,9R)-3-oxo-alpha-Ionyl-9-O-alpha-rhamnopyranosyl-(1''-6')-beta-glucopyranoside | MeSH |
|
|---|
| Chemical Formula | C25H40O11 |
|---|
| Average Molecular Mass | 516.579 g/mol |
|---|
| Monoisotopic Mass | 516.257 g/mol |
|---|
| CAS Registry Number | 351412-97-0 |
|---|
| IUPAC Name | 3,5,5-trimethyl-4-[(1E)-3-[(3,4,5-trihydroxy-6-{[(3,4,5-trihydroxy-6-methyloxan-2-yl)oxy]methyl}oxan-2-yl)oxy]but-1-en-1-yl]cyclohex-2-en-1-one |
|---|
| Traditional Name | 3,5,5-trimethyl-4-[(1E)-3-[(3,4,5-trihydroxy-6-{[(3,4,5-trihydroxy-6-methyloxan-2-yl)oxy]methyl}oxan-2-yl)oxy]but-1-en-1-yl]cyclohex-2-en-1-one |
|---|
| SMILES | CC(OC1OC(COC2OC(C)C(O)C(O)C2O)C(O)C(O)C1O)\C=C\C1C(C)=CC(=O)CC1(C)C |
|---|
| InChI Identifier | InChI=1S/C25H40O11/c1-11-8-14(26)9-25(4,5)15(11)7-6-12(2)34-24-22(32)20(30)18(28)16(36-24)10-33-23-21(31)19(29)17(27)13(3)35-23/h6-8,12-13,15-24,27-32H,9-10H2,1-5H3/b7-6+ |
|---|
| InChI Key | AFWVBXLXFDAISA-VOTSOKGWSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as fatty acyl glycosides of mono- and disaccharides. Fatty acyl glycosides of mono- and disaccharides are compounds composed of a mono- or disaccharide moiety linked to one hydroxyl group of a fatty alcohol or of a phosphorylated alcohol (phosphoprenols), a hydroxy fatty acid or to one carboxyl group of a fatty acid (ester linkage) or to an amino alcohol. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Fatty Acyls |
|---|
| Sub Class | Fatty acyl glycosides |
|---|
| Direct Parent | Fatty acyl glycosides of mono- and disaccharides |
|---|
| Alternative Parents | |
|---|
| Substituents | - Fatty acyl glycoside of mono- or disaccharide
- Cyclofarsesane sesquiterpenoid
- Megastigmane sesquiterpenoid
- Sesquiterpenoid
- Ionone derivative
- Alkyl glycoside
- Disaccharide
- Glycosyl compound
- O-glycosyl compound
- Cyclohexenone
- Oxane
- Ketone
- Cyclic ketone
- Secondary alcohol
- Organoheterocyclic compound
- Acetal
- Oxacycle
- Polyol
- Organooxygen compound
- Hydrocarbon derivative
- Organic oxide
- Carbonyl group
- Organic oxygen compound
- Alcohol
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0002-8563920000-6d637f3bb63294d13376 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-0007-5745109000-6c335ef6d1ac62376a06 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0aov-1973540000-1d92cdf615f5cbc14d55 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a4l-1941000000-41b68d5cf0ccd415e677 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-052f-3920000000-7bd009bf850e9c34fbcc | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-066r-4793260000-b4cad6d21bda656ad3bf | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4i-3892100000-a9892da6426a05132d65 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-5890000000-457cbcafc002abac80af | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-052f-0911310000-87d9d2f0f53f56452b50 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0uds-1900300000-233b91debba76cff1cab | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0007-3911000000-d9ce531ce66adabe4d5a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0191240000-a41d28e391146d3f094c | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-05n0-4914510000-dd418fb4e1ecc04e8689 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4l-8920000000-e3b8bc49235e3225aa05 | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0038029 |
|---|
| FooDB ID | FDB017239 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | C00045921 |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | Not Available |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 131752277 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|