| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 02:30:26 UTC |
|---|
| Update Date | 2016-11-09 01:19:13 UTC |
|---|
| Accession Number | CHEM031246 |
|---|
| Identification |
|---|
| Common Name | Annuolide B |
|---|
| Class | Small Molecule |
|---|
| Description | Annuolide B is found in fats and oils. Annuolide B is a constituent of Helianthus annuus (sunflower). |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
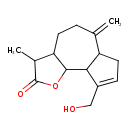
| Chemical Structure | |
|---|
| Synonyms | Not Available |
|---|
| Chemical Formula | C15H20O3 |
|---|
| Average Molecular Mass | 248.318 g/mol |
|---|
| Monoisotopic Mass | 248.141 g/mol |
|---|
| CAS Registry Number | 152442-50-7 |
|---|
| IUPAC Name | 9-(hydroxymethyl)-3-methyl-6-methylidene-2H,3H,3aH,4H,5H,6H,6aH,7H,9aH,9bH-azuleno[4,5-b]furan-2-one |
|---|
| Traditional Name | 9-(hydroxymethyl)-3-methyl-6-methylidene-3H,3aH,4H,5H,6aH,7H,9aH,9bH-azuleno[4,5-b]furan-2-one |
|---|
| SMILES | CC1C2CCC(=C)C3CC=C(CO)C3C2OC1=O |
|---|
| InChI Identifier | InChI=1S/C15H20O3/c1-8-3-5-12-9(2)15(17)18-14(12)13-10(7-16)4-6-11(8)13/h4,9,11-14,16H,1,3,5-7H2,2H3 |
|---|
| InChI Key | PTSCCEBQBSUFRX-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as guaianolides and derivatives. These are diterpene lactones with a structure characterized by the presence of a gamma-lactone fused to a guaiane, forming 3,6,9-trimethyl-azuleno[4,5-b]furan-2-one or a derivative. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Terpene lactones |
|---|
| Direct Parent | Guaianolides and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Guaianolide-skeleton
- Guaiane sesquiterpenoid
- Sesquiterpenoid
- Gamma butyrolactone
- Tetrahydrofuran
- Carboxylic acid ester
- Lactone
- Monocarboxylic acid or derivatives
- Carboxylic acid derivative
- Oxacycle
- Organoheterocyclic compound
- Primary alcohol
- Organooxygen compound
- Hydrocarbon derivative
- Organic oxide
- Organic oxygen compound
- Alcohol
- Carbonyl group
- Aliphatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aliphatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0fi0-4940000000-3486fb53f5f05cc3b35d | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-0c00-2390000000-e086fbbf8b870ffbb7eb | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000t-0290000000-b171c02b5d5b79fe45fc | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-053r-0950000000-d0346f6519713a6a83fa | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0pb9-5910000000-6edf2b70fd31a9fcd0e8 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-0190000000-3be64209b08390596a85 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0ftb-0290000000-df42236a307a1cbf1a70 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0udi-5910000000-d36c552c0647248e59bb | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-0090000000-7b3a58b796a0e4263959 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0002-0090000000-c1c969788f7ea66e674a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-066s-5940000000-0fd0704b5a1d3b133bd1 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0002-0090000000-d075ce931df6d8d6bcbb | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0002-0490000000-fdb3cd9d49cac2fdb675 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-002g-3920000000-607fbc8b18a580ca8560 | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0037800 |
|---|
| FooDB ID | FDB016944 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 35014474 |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 131752239 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|