| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 02:30:13 UTC |
|---|
| Update Date | 2016-11-09 01:19:13 UTC |
|---|
| Accession Number | CHEM031240 |
|---|
| Identification |
|---|
| Common Name | (3S,3'R,5R,6R)-7',8'-Didehydro-3,6-epoxy-5,6-dihydro-beta,beta-carotene-3',5-diol |
|---|
| Class | Small Molecule |
|---|
| Description | (3S,3'R,5R,6R)-7',8'-Didehydro-3,6-epoxy-5,6-dihydro-beta,beta-carotene-3',5-diol is found in mollusks. (3S,3'R,5R,6R)-7',8'-Didehydro-3,6-epoxy-5,6-dihydro-beta,beta-carotene-3',5-diol is a constituent of the oyster Crassostrea gigas. |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
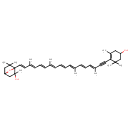
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (3S,3'r,5R,6R)-7',8'-Didehydro-3,6-epoxy-5,6-dihydro-b,b-carotene-3',5-diol | Generator | | (3S,3'r,5R,6R)-7',8'-Didehydro-3,6-epoxy-5,6-dihydro-β,β-carotene-3',5-diol | Generator | | Diatoxanthin 3,6-epoxide | HMDB |
|
|---|
| Chemical Formula | C40H54O3 |
|---|
| Average Molecular Mass | 582.855 g/mol |
|---|
| Monoisotopic Mass | 582.407 g/mol |
|---|
| CAS Registry Number | 256505-52-9 |
|---|
| IUPAC Name | 1-[(1E,3E,5E,7E,9E,11E,13E,15E)-18-(4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl)-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15-octaen-17-yn-1-yl]-2,6,6-trimethyl-7-oxabicyclo[2.2.1]heptan-2-ol |
|---|
| Traditional Name | 1-[(1E,3E,5E,7E,9E,11E,13E,15E)-18-(4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl)-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15-octaen-17-yn-1-yl]-2,6,6-trimethyl-7-oxabicyclo[2.2.1]heptan-2-ol |
|---|
| SMILES | C\C(\C=C\C=C(/C)\C=C\C12OC(CC1(C)C)CC2(C)O)=C/C=C/C=C(\C)/C=C/C=C(\C)C#CC1=C(C)CC(O)CC1(C)C |
|---|
| InChI Identifier | InChI=1S/C40H54O3/c1-29(17-13-19-31(3)21-22-36-33(5)25-34(41)26-37(36,6)7)15-11-12-16-30(2)18-14-20-32(4)23-24-40-38(8,9)27-35(43-40)28-39(40,10)42/h11-20,23-24,34-35,41-42H,25-28H2,1-10H3/b12-11+,17-13+,18-14+,24-23+,29-15+,30-16+,31-19+,32-20+ |
|---|
| InChI Key | QOCBLJWPUILPNE-RQCOEWNJSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as triterpenoids. These are terpene molecules containing six isoprene units. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Triterpenoids |
|---|
| Direct Parent | Triterpenoids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Triterpenoid
- Monosaccharide
- Tetrahydrofuran
- Tertiary alcohol
- Cyclic alcohol
- Secondary alcohol
- Oxacycle
- Organoheterocyclic compound
- Ether
- Dialkyl ether
- Organic oxygen compound
- Hydrocarbon derivative
- Organooxygen compound
- Alcohol
- Aliphatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aliphatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-014r-2000090000-e42eb3a703c7e03331ee | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-000i-3000049000-bdb4c4893b6313201266 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS ("(3S,3'R,5R,6R)-7',8'-Didehydro-3,6-epoxy-5,6-dihydro-beta,beta-carotene-3',5-diol,1TMS,#1" TMS) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_1) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_2) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_1) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0159-0121390000-e924c6073c91ceed028d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-01pk-0795440000-2f6c56f95051facdfc6a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0o6s-5895200000-c6cdcfe90bf3c50c91d8 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-001i-0000090000-077c3df367133f1b7bb1 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-01q9-0100090000-dec6e920e0b2f7dd7436 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0l0i-2900260000-164e702e22adac48ec94 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-001i-0102090000-db8b9989cc2ea1f1c3f1 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-001i-0214290000-1fa451ca95cf91dd670b | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0abj-4319040000-91be36786ece9ae635e0 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-001i-0102090000-734e2127cee4490a75d2 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a4r-6354390000-8ca12bce6232a357aeda | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-01r2-1219100000-543a464e2e1846fa2c60 | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0037794 |
|---|
| FooDB ID | FDB016938 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | C00023238 |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 35014471 |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 131752235 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|