| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 02:27:19 UTC |
|---|
| Update Date | 2016-11-09 01:19:13 UTC |
|---|
| Accession Number | CHEM031183 |
|---|
| Identification |
|---|
| Common Name | (+)-1(9),10-Pacifigorgiadiene |
|---|
| Class | Small Molecule |
|---|
| Description | (+)-1(9),10-Pacifigorgiadiene is found in fats and oils. (+)-1(9),10-Pacifigorgiadiene is a constituent of Valeriana officinalis (valerian). |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
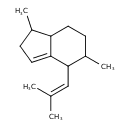
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (-)-Pacifigorgia-1(9),10-diene | HMDB |
|
|---|
| Chemical Formula | C15H24 |
|---|
| Average Molecular Mass | 204.351 g/mol |
|---|
| Monoisotopic Mass | 204.188 g/mol |
|---|
| CAS Registry Number | 351222-62-3 |
|---|
| IUPAC Name | 1,5-dimethyl-4-(2-methylprop-1-en-1-yl)-2,4,5,6,7,7a-hexahydro-1H-indene |
|---|
| Traditional Name | 1,5-dimethyl-4-(2-methylprop-1-en-1-yl)-2,4,5,6,7,7a-hexahydro-1H-indene |
|---|
| SMILES | CC1CC=C2C1CCC(C)C2C=C(C)C |
|---|
| InChI Identifier | InChI=1S/C15H24/c1-10(2)9-15-12(4)5-7-13-11(3)6-8-14(13)15/h8-9,11-13,15H,5-7H2,1-4H3 |
|---|
| InChI Key | DMSFMSTWAVYMQU-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as bicyclic monoterpenoids. These are monoterpenoids containing exactly 2 rings, which are fused to each other. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Monoterpenoids |
|---|
| Direct Parent | Bicyclic monoterpenoids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Bicyclic monoterpenoid
- Branched unsaturated hydrocarbon
- Polycyclic hydrocarbon
- Cyclic olefin
- Unsaturated aliphatic hydrocarbon
- Unsaturated hydrocarbon
- Olefin
- Hydrocarbon
- Aliphatic homopolycyclic compound
|
|---|
| Molecular Framework | Aliphatic homopolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-01tj-2900000000-60a942327f4d3070c504 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0a4i-1590000000-1f80a66a73a7bd3a9d13 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0cdj-4920000000-2888abbc7e719f84919b | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0gbc-9500000000-04db59ec459ceb3233b5 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0udi-0090000000-4e4a2bf9614203ba3df5 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0udi-0290000000-de99d4c72ff08b7b36cb | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-052s-3900000000-608844988f80e368c487 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0udi-0090000000-7ccf03fa1149a1e9f55f | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0udi-0090000000-7ccf03fa1149a1e9f55f | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0zms-1920000000-c476f0cada7a9d619610 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0a4i-0390000000-deb4ddfb98304c13b382 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0bta-5940000000-10124c9e05bb2d329348 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-052f-6900000000-a996f9c0288432e98535 | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0037677 |
|---|
| FooDB ID | FDB016804 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | C00036286 |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 35014457 |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 131752220 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|