| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 02:24:37 UTC |
|---|
| Update Date | 2016-11-09 01:19:12 UTC |
|---|
| Accession Number | CHEM031126 |
|---|
| Identification |
|---|
| Common Name | (10betaH,11xi)-11-Hydroxy-13-nor-6-eremophilen-8-one |
|---|
| Class | Small Molecule |
|---|
| Description | (10betaH,11xi)-11-Hydroxy-13-nor-6-eremophilen-8-one is found in green vegetables. (10betaH,11xi)-11-Hydroxy-13-nor-6-eremophilen-8-one is a constituent of Petasites japonicus ssp. giganteus (Japanese butterbur). |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
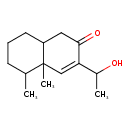
| Chemical Structure | |
|---|
| Synonyms | Not Available |
|---|
| Chemical Formula | C14H22O2 |
|---|
| Average Molecular Mass | 222.323 g/mol |
|---|
| Monoisotopic Mass | 222.162 g/mol |
|---|
| CAS Registry Number | 348615-40-7 |
|---|
| IUPAC Name | 3-(1-hydroxyethyl)-4a,5-dimethyl-1,2,4a,5,6,7,8,8a-octahydronaphthalen-2-one |
|---|
| Traditional Name | 3-(1-hydroxyethyl)-4a,5-dimethyl-1,5,6,7,8,8a-hexahydronaphthalen-2-one |
|---|
| SMILES | CC(O)C1=CC2(C)C(C)CCCC2CC1=O |
|---|
| InChI Identifier | InChI=1S/C14H22O2/c1-9-5-4-6-11-7-13(16)12(10(2)15)8-14(9,11)3/h8-11,15H,4-7H2,1-3H3 |
|---|
| InChI Key | YAQFZWZUAZKHAL-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as eremophilane, 8,9-secoeremophilane and furoeremophilane sesquiterpenoids. These are sesquiterpenoids with a structure based either on the eremophilane skeleton, its 8,9-seco derivative, or the furoeremophilane skeleton. Eremophilanes have been shown to be derived from eudesmanes by migration of the methyl group at C-10 to C-5. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Sesquiterpenoids |
|---|
| Direct Parent | Eremophilane, 8,9-secoeremophilane and furoeremophilane sesquiterpenoids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Eremophilane sesquiterpenoid
- Cyclohexenone
- Cyclic ketone
- Secondary alcohol
- Ketone
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Carbonyl group
- Alcohol
- Aliphatic homopolycyclic compound
|
|---|
| Molecular Framework | Aliphatic homopolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0a4i-0930000000-41592ff1f3ceed0e9a94 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-00b9-6390000000-a08b41e5e4895ea6556b | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0ab9-0190000000-226a983c8da16a97b25e | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0abi-2950000000-f7f2fe74a005fc90bad5 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0ftu-5900000000-4ca6ab10df9b6634a8cf | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00di-0190000000-0c4c20d02e816763630d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00fr-0590000000-3bc98c5ff6a775285378 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-01tc-3900000000-3274a540855b465506eb | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00di-0090000000-277654a816c7fdb457aa | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00di-1190000000-c202297897ca3d1b3eab | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0hfx-2920000000-c9cccabb21e3b7302368 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00di-0090000000-7a435785fe23f6a458f1 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0ab9-0960000000-84930377b62fffa4d469 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0007-9600000000-8eee48bd6c2a7c8e2bfe | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0037605 |
|---|
| FooDB ID | FDB016718 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 35014443 |
|---|
| ChEBI ID | 174141 |
|---|
| PubChem Compound ID | 85186102 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|