| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 02:09:31 UTC |
|---|
| Update Date | 2016-11-09 01:19:08 UTC |
|---|
| Accession Number | CHEM030778 |
|---|
| Identification |
|---|
| Common Name | Quassinol |
|---|
| Class | Small Molecule |
|---|
| Description | Constituent of Quassia amara (Surinam quassia) |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Bisnorquassin | HMDB |
|
|---|
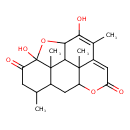
| Chemical Formula | C20H24O6 |
|---|
| Average Molecular Mass | 360.401 g/mol |
|---|
| Monoisotopic Mass | 360.157 g/mol |
|---|
| CAS Registry Number | 21018-87-1 |
|---|
| IUPAC Name | 5,8-dihydroxy-2,9,15,17-tetramethyl-6,13-dioxapentacyclo[12.3.1.0⁵,¹⁷.0⁷,¹⁶.0¹⁰,¹⁵]octadeca-8,10-diene-4,12-dione |
|---|
| Traditional Name | 5,8-dihydroxy-2,9,15,17-tetramethyl-6,13-dioxapentacyclo[12.3.1.0⁵,¹⁷.0⁷,¹⁶.0¹⁰,¹⁵]octadeca-8,10-diene-4,12-dione |
|---|
| SMILES | CC1CC(=O)C2(O)OC3C4C2(C)C1CC1OC(=O)C=C(C(C)=C3O)C41C |
|---|
| InChI Identifier | InChI=1S/C20H24O6/c1-8-5-12(21)20(24)19(4)10(8)6-13-18(3)11(7-14(22)25-13)9(2)15(23)16(26-20)17(18)19/h7-8,10,13,16-17,23-24H,5-6H2,1-4H3 |
|---|
| InChI Key | WKNPIBKVHHVICI-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as quassinoids. These are a group of compounds chemically degraded from triterpenes. According to their basic skeleton, quassinoids are categorized into five distinct groups, C-18, C-19, C-20, C-22 and C-25 types. The C-20 quassinoids can be further classified into two types, tetracyclic and the pentacyclic. The tetracyclic variety does not have oxygenation at C-20, while the pentacyclic quassinoids possess additional oxygenation at C-20 that allows for the formation of an additional ring. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Terpene lactones |
|---|
| Direct Parent | Quassinoids |
|---|
| Alternative Parents | |
|---|
| Substituents | - C-20 quassinoid skeleton
- Quassinoid
- Naphthopyran
- Naphthalene
- Dihydropyranone
- Pyran
- Cyclic alcohol
- Alpha,beta-unsaturated carboxylic ester
- Tetrahydrofuran
- Enoate ester
- Lactone
- Ketone
- Hemiacetal
- Carboxylic acid ester
- Oxacycle
- Monocarboxylic acid or derivatives
- Enol
- Organoheterocyclic compound
- Carboxylic acid derivative
- Organooxygen compound
- Hydrocarbon derivative
- Organic oxide
- Organic oxygen compound
- Carbonyl group
- Aliphatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aliphatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-00kf-9005000000-acaa165a121140aa7d49 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-010c-9000700000-0042a653c5daba1bbfae | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0ik9-0009000000-18a56bd4b5f912a7ebe9 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0wmi-0019000000-6ff9522e692eb16b60e1 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0006-9023000000-d5fcc0c203860d251663 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0aor-0009000000-cc9cc9fe9fc7faa50f83 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0aor-0009000000-5e6b0fe8b634b0fad5fb | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-066v-6094000000-925320bfb648d5fe4d68 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0009000000-d6fe99be8012d24cd96c | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4i-0009000000-02d8dcc6f2e6225173f2 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-0009000000-2bf57022a871e5d8c38f | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03di-0009000000-6072fd31342288e0ad83 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03dl-0019000000-e7b1c2ea89e1171d2075 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03di-0019000000-32977a6392faca67f3c7 | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0037235 |
|---|
| FooDB ID | FDB016242 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | Not Available |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 131752166 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|