| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 02:04:59 UTC |
|---|
| Update Date | 2016-11-09 01:19:07 UTC |
|---|
| Accession Number | CHEM030685 |
|---|
| Identification |
|---|
| Common Name | Sporotrichiol |
|---|
| Class | Small Molecule |
|---|
| Description | Sporotrichiol is produced by Fusarium sporotrichioides. |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
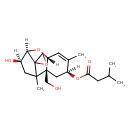
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 12,13-Epoxy-9-trichothecene-3,8,15-triol 8-(3-methylbutenoate), 9ci | HMDB | | (2'r,4's,7'r,9'r,10'r)-10'-Hydroxy-2'-(hydroxymethyl)-1',5'-dimethyl-8'-oxaspiro[oxirane-2,12'-tricyclo[7.2.1.0²,⁷]dodecan]-5'-en-4'-yl 3-methylbutanoic acid | Generator |
|
|---|
| Chemical Formula | C20H30O6 |
|---|
| Average Molecular Mass | 366.449 g/mol |
|---|
| Monoisotopic Mass | 366.204 g/mol |
|---|
| CAS Registry Number | 101401-89-2 |
|---|
| IUPAC Name | (2'R,4'S,7'R,9'R,10'R)-10'-hydroxy-2'-(hydroxymethyl)-1',5'-dimethyl-8'-oxaspiro[oxirane-2,12'-tricyclo[7.2.1.0²,⁷]dodecan]-5'-en-4'-yl 3-methylbutanoate |
|---|
| Traditional Name | (2'R,4'S,7'R,9'R,10'R)-10'-hydroxy-2'-(hydroxymethyl)-1',5'-dimethyl-8'-oxaspiro[oxirane-2,12'-tricyclo[7.2.1.0²,⁷]dodecan]-5'-en-4'-yl 3-methylbutanoate |
|---|
| SMILES | [H][C@@]12O[C@]3([H])C=C(C)[C@H](C[C@]3(CO)C(C)(C[C@H]1O)C21CO1)OC(=O)CC(C)C |
|---|
| InChI Identifier | InChI=1S/C20H30O6/c1-11(2)5-16(23)25-14-8-19(9-21)15(6-12(14)3)26-17-13(22)7-18(19,4)20(17)10-24-20/h6,11,13-15,17,21-22H,5,7-10H2,1-4H3/t13-,14+,15-,17-,18?,19-,20?/m1/s1 |
|---|
| InChI Key | RCFUVEKOPPKTBN-UBOCPNNGSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as trichothecenes. These are sesquiterpene mycotoxins structurally characterized by the presence of an epoxide ring and a benzopyran derivative with a variant number of hydroxyl, acetyl, or other substituents. The most important structural features causing the biological activities of trichothecenes are the 12,13-epoxy ring, the presence of hydroxyl or acetyl groups at appropriate positions on the trichothecene nucleus and the structure and position of the side-chain. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Sesquiterpenoids |
|---|
| Direct Parent | Trichothecenes |
|---|
| Alternative Parents | |
|---|
| Substituents | - Trichothecene skeleton
- Fatty acid ester
- Oxepane
- Oxane
- Fatty acyl
- Cyclic alcohol
- Carboxylic acid ester
- Secondary alcohol
- Organoheterocyclic compound
- Carboxylic acid derivative
- Oxacycle
- Dialkyl ether
- Oxirane
- Ether
- Monocarboxylic acid or derivatives
- Carbonyl group
- Alcohol
- Organic oxygen compound
- Organooxygen compound
- Organic oxide
- Hydrocarbon derivative
- Primary alcohol
- Aliphatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aliphatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0079-6497000000-1ddedc1b057e77bd4575 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-0a4j-9233500000-7b669322a885bca62cbf | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014j-3049000000-8a88aac36a874cbc59e9 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00kg-9263000000-fcbe256f7aca0cc33545 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-05mo-9150000000-489378a452ddb02fbf10 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-014i-1139000000-530a7c08b133d87b1265 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0gx0-3497000000-debd9c54ef1e2150dfd0 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-052f-4910000000-c80fc018ec2b0a5c2b91 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-014i-0119000000-1d0db51fdb87d5161175 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-001i-9281000000-2df46ae6e3fdc97824b9 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00lu-9300000000-f8b9e491cd318b6c72fc | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014i-0096000000-ae594018af1101c21e4d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-014j-5192000000-9351d59508410d5fdc25 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0aou-9110000000-26bed465f1f5c380dbe4 | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0037062 |
|---|
| FooDB ID | FDB016045 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | C00012650 |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 26503953 |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 131752137 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|