| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 01:59:44 UTC |
|---|
| Update Date | 2016-11-09 01:19:06 UTC |
|---|
| Accession Number | CHEM030569 |
|---|
| Identification |
|---|
| Common Name | 7,8-Dihydroparasiloxanthin |
|---|
| Class | Small Molecule |
|---|
| Description | 7,8-Dihydroparasiloxanthin is found in fishes. 7,8-Dihydroparasiloxanthin is a constituent of skin and fin of Japanese catfish (Parasilurus asotus). |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
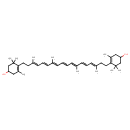
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 7,7',8,8'-tetrahydro-b,b-Carotene-3,3'-diol | HMDB | | 7,7',8,8'-tetrahydro-beta,Beat-carotene-3,3'-diol | HMDB |
|
|---|
| Chemical Formula | C40H60O2 |
|---|
| Average Molecular Mass | 572.903 g/mol |
|---|
| Monoisotopic Mass | 572.459 g/mol |
|---|
| CAS Registry Number | 62994-71-2 |
|---|
| IUPAC Name | 4-[(3E,5E,7E,9E,11E,13E,15E)-18-(4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl)-3,7,12,16-tetramethyloctadeca-3,5,7,9,11,13,15-heptaen-1-yl]-3,5,5-trimethylcyclohex-3-en-1-ol |
|---|
| Traditional Name | 4-[(3E,5E,7E,9E,11E,13E,15E)-18-(4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl)-3,7,12,16-tetramethyloctadeca-3,5,7,9,11,13,15-heptaen-1-yl]-3,5,5-trimethylcyclohex-3-en-1-ol |
|---|
| SMILES | C\C(CCC1=C(C)CC(O)CC1(C)C)=C/C=C/C(/C)=C/C=C/C=C(\C)/C=C/C=C(\C)CCC1=C(C)CC(O)CC1(C)C |
|---|
| InChI Identifier | InChI=1S/C40H60O2/c1-29(17-13-19-31(3)21-23-37-33(5)25-35(41)27-39(37,7)8)15-11-12-16-30(2)18-14-20-32(4)22-24-38-34(6)26-36(42)28-40(38,9)10/h11-20,35-36,41-42H,21-28H2,1-10H3/b12-11+,17-13+,18-14+,29-15+,30-16+,31-19+,32-20+ |
|---|
| InChI Key | DCNLLBVWQNTIMR-PJQROKOUSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as xanthophylls. These are carotenoids containing an oxygenated carotene backbone. Carotenes are characterized by the presence of two end-groups (mostly cyclohexene rings, but also cyclopentene rings or acyclic groups) linked by a long branched alkyl chain. Carotenes belonging form a subgroup of the carotenoids family. Xanthophylls arise by oxygenation of the carotene backbone. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Tetraterpenoids |
|---|
| Direct Parent | Xanthophylls |
|---|
| Alternative Parents | |
|---|
| Substituents | - Xanthophyll
- Secondary alcohol
- Organic oxygen compound
- Hydrocarbon derivative
- Organooxygen compound
- Alcohol
- Aliphatic homomonocyclic compound
|
|---|
| Molecular Framework | Aliphatic homomonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0pvi-2400590000-c11af1ce91f168d82e1b | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-0059-2310419000-1d8059bf4bd8e8b0f91a | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS ("7,8-Dihydroparasiloxanthin,1TMS,#1" TMS) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_1) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_1) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-05fr-0111090000-2cc51f3fcba51a1f72c5 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0uy0-0898560000-ab61da7f0d3e75663c5d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0f81-0669340000-c865c573b984730349fb | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00di-0000090000-83e5106d1d500627a49c | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00di-0000090000-d3e5aee2699a8a6b9af5 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0ap3-0453390000-a5731de82e7c94cfa49c | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00di-0100090000-d182b1aeafa7f7d31bc0 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00dr-0512190000-fc89f2d0ece35da077f8 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0fb9-0219500000-56ca5697b4489e91eedb | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-05g0-0152690000-32ecd46bccf8b3860e31 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-0643970000-aa5a1a47aba5ed7f4475 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0ugr-0593200000-975dc68286bff4a15889 | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0036916 |
|---|
| FooDB ID | FDB015881 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | C00022867 |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 35014314 |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 87442432 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|