| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 01:57:32 UTC |
|---|
| Update Date | 2016-11-09 01:19:05 UTC |
|---|
| Accession Number | CHEM030527 |
|---|
| Identification |
|---|
| Common Name | Alloxanthin |
|---|
| Class | Small Molecule |
|---|
| Description | Alloxanthin is found in channel catfish. Alloxanthin is a constituent of many shellfish including the giant scallop (Pecten maximus) and edible mussel (Mytilus edulis). |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
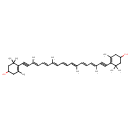
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 7,7',8,8'-tetradehydro-b,b-Carotene-3,3'-diol | HMDB | | Cryptomonaxanthin | HMDB | | Manixanthin | HMDB | | Pectenoxanthin | HMDB | | Alloxanthin | MeSH |
|
|---|
| Chemical Formula | C40H52O2 |
|---|
| Average Molecular Mass | 564.840 g/mol |
|---|
| Monoisotopic Mass | 564.397 g/mol |
|---|
| CAS Registry Number | 28380-31-6 |
|---|
| IUPAC Name | 4-[(3E,5E,7E,9E,11E,13E,15E)-18-(4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl)-3,7,12,16-tetramethyloctadeca-3,5,7,9,11,13,15-heptaen-1,17-diyn-1-yl]-3,5,5-trimethylcyclohex-3-en-1-ol |
|---|
| Traditional Name | 4-[(3E,5E,7E,9E,11E,13E,15E)-18-(4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl)-3,7,12,16-tetramethyloctadeca-3,5,7,9,11,13,15-heptaen-1,17-diyn-1-yl]-3,5,5-trimethylcyclohex-3-en-1-ol |
|---|
| SMILES | C\C(\C=C\C=C(/C)C#CC1=C(C)CC(O)CC1(C)C)=C/C=C/C=C(\C)/C=C/C=C(\C)C#CC1=C(C)CC(O)CC1(C)C |
|---|
| InChI Identifier | InChI=1S/C40H52O2/c1-29(17-13-19-31(3)21-23-37-33(5)25-35(41)27-39(37,7)8)15-11-12-16-30(2)18-14-20-32(4)22-24-38-34(6)26-36(42)28-40(38,9)10/h11-20,35-36,41-42H,25-28H2,1-10H3/b12-11+,17-13+,18-14+,29-15+,30-16+,31-19+,32-20+ |
|---|
| InChI Key | DVICWXUADSCSLL-PJQROKOUSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as diterpenoids. These are terpene compounds formed by four isoprene units. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Diterpenoids |
|---|
| Direct Parent | Diterpenoids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Diterpenoid
- Secondary alcohol
- Organic oxygen compound
- Hydrocarbon derivative
- Organooxygen compound
- Alcohol
- Aliphatic homomonocyclic compound
|
|---|
| Molecular Framework | Aliphatic homomonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0002-2000190000-f6102c1c421c7d41666e | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-00di-2100049000-676bc1713d2a18308c8c | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS ("Alloxanthin,1TMS,#1" TMS) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_1) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_1) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014j-0011090000-8c758f03201e45452f6c | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0002-0759450000-382bf4f291a09351585a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0002-1439430000-2f339e64328fce3ff30a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03di-0000090000-5d370f6307a42c62a5c2 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-03di-0000090000-64575b0b1571dc9dceec | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0002-0312290000-598f440dab1a11474bbf | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03di-0002090000-37297b1ee91dfcd3cf82 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-03di-0016090000-8241f0bb8d4fc69a2649 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-001j-0339210000-6a77821b58aa9bb0fc90 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014i-0130390000-e47bef973f43bacd2e35 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0092-0248490000-041e636a041dc9b760d5 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0bvi-0029500000-df4e73a776dd015c521a | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0036874 |
|---|
| FooDB ID | FDB015830 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | C00022862 |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 10469527 |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 12305997 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|