| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 01:52:26 UTC |
|---|
| Update Date | 2016-11-09 01:19:04 UTC |
|---|
| Accession Number | CHEM030407 |
|---|
| Identification |
|---|
| Common Name | ent-7-Oxo-8(14),15-pimaradien-19-oic acid |
|---|
| Class | Small Molecule |
|---|
| Description | ent-7-Oxo-8(14),15-pimaradien-19-oic acid is found in green vegetables. ent-7-Oxo-8(14),15-pimaradien-19-oic acid is a constituent of Aralia cordata (udo). |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
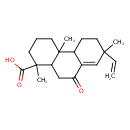
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| ent-7-oxo-8(14),15-Pimaradien-19-Oate | Generator | | 7-Ethenyl-1,4a,7-trimethyl-9-oxo-1,2,3,4,4a,4b,5,6,7,9,10,10a-dodecahydrophenanthrene-1-carboxylate | HMDB |
|
|---|
| Chemical Formula | C20H28O3 |
|---|
| Average Molecular Mass | 316.435 g/mol |
|---|
| Monoisotopic Mass | 316.204 g/mol |
|---|
| CAS Registry Number | 23807-90-1 |
|---|
| IUPAC Name | 7-ethenyl-1,4a,7-trimethyl-9-oxo-1,2,3,4,4a,4b,5,6,7,9,10,10a-dodecahydrophenanthrene-1-carboxylic acid |
|---|
| Traditional Name | 7-ethenyl-1,4a,7-trimethyl-9-oxo-2,3,4,4b,5,6,10,10a-octahydrophenanthrene-1-carboxylic acid |
|---|
| SMILES | CC1(CCC2C(=C1)C(=O)CC1C2(C)CCCC1(C)C(O)=O)C=C |
|---|
| InChI Identifier | InChI=1S/C20H28O3/c1-5-18(2)10-7-14-13(12-18)15(21)11-16-19(14,3)8-6-9-20(16,4)17(22)23/h5,12,14,16H,1,6-11H2,2-4H3,(H,22,23) |
|---|
| InChI Key | TWQIAJPCUCIDQX-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as diterpenoids. These are terpene compounds formed by four isoprene units. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Diterpenoids |
|---|
| Direct Parent | Diterpenoids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Pimarane diterpenoid
- Diterpenoid
- Phenanthrene
- Hydrophenanthrene
- Ketone
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Carboxylic acid derivative
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Carbonyl group
- Aliphatic homopolycyclic compound
|
|---|
| Molecular Framework | Aliphatic homopolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0udi-0794000000-ddc96929ca6d5c0e39d8 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-00di-6269000000-5b9091cdf94ef95f2a79 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00kb-1095000000-ac57c16d572c3305b7ef | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0fka-5291000000-b1d069c6f0525aecc9e2 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0udi-9120000000-4ad13e9bd13a12479aec | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-014i-0059000000-e8eacf55277da6db08c3 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00xr-0093000000-6342531ae2ff7f849719 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-052e-2090000000-aa72dc5d503a40bd5b70 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-014i-0009000000-d337ec43015bf5d00be6 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-014i-0009000000-d3502f75eded219ab375 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-03y3-1194000000-49502f69ba5828b8688f | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014i-0095000000-5245137f70ceaa4fa26d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00xv-0290000000-9766a1e0342a3d913fd3 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-06dr-7980000000-1d476ef65f0e0ec0b4cb | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0036747 |
|---|
| FooDB ID | FDB015685 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | C00057278 |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 19576089 |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 20744631 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|