| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 01:52:02 UTC |
|---|
| Update Date | 2016-11-09 01:19:04 UTC |
|---|
| Accession Number | CHEM030396 |
|---|
| Identification |
|---|
| Common Name | Bilobalide A |
|---|
| Class | Small Molecule |
|---|
| Description | Constituent of leaves of Ginkgo biloba (ginkgo). Bilobalide A is found in ginkgo nuts and fats and oils. |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
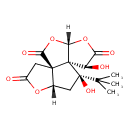
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Bilobalide | HMDB | | (-)-Bilobalide | MeSH |
|
|---|
| Chemical Formula | C15H18O8 |
|---|
| Average Molecular Mass | 326.299 g/mol |
|---|
| Monoisotopic Mass | 326.100 g/mol |
|---|
| CAS Registry Number | 33570-04-6 |
|---|
| IUPAC Name | (1S,4R,7S,8S,9R,11S)-9-tert-butyl-7,9-dihydroxy-3,5,12-trioxatetracyclo[6.6.0.0¹,¹¹.0⁴,⁸]tetradecane-2,6,13-trione |
|---|
| Traditional Name | (1S,4R,7S,8S,9R,11S)-9-tert-butyl-7,9-dihydroxy-3,5,12-trioxatetracyclo[6.6.0.0¹,¹¹.0⁴,⁸]tetradecane-2,6,13-trione |
|---|
| SMILES | [H][C@]12C[C@@](O)(C(C)(C)C)[C@]34[C@H](O)C(=O)O[C@@]3([H])OC(=O)[C@]14CC(=O)O2 |
|---|
| InChI Identifier | InChI=1S/C15H18O8/c1-12(2,3)14(20)4-6-13(5-7(16)21-6)10(19)23-11-15(13,14)8(17)9(18)22-11/h6,8,11,17,20H,4-5H2,1-3H3/t6-,8+,11-,13-,14+,15+/m0/s1 |
|---|
| InChI Key | MOLPUWBMSBJXER-NRSGSQFTSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as ginkgolides and bilobalides. These are diterpene lactones with a structure based either on the gingkolide or the bilobalide skeleton. The ginkgolide skeleton is a very rigid structure consisting of hexacyclic C20 trilactone. The cis-fused F/A/D/C ring junction forms an empty semi-ball hole, the D ring contains a cage form tetrahydrofuran ring which occupies the center of the empty hole, and the oxygen atoms of the D,C and F ring and 10-hydroxyl group consist of an analogous crown ether structure. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Terpene lactones |
|---|
| Direct Parent | Ginkgolides and bilobalides |
|---|
| Alternative Parents | |
|---|
| Substituents | - Bilobalide
- Diterpenoid
- Tricarboxylic acid or derivatives
- Furofuran
- Acylal
- Gamma butyrolactone
- Cyclic alcohol
- Tetrahydrofuran
- Tertiary alcohol
- Carboxylic acid ester
- Lactone
- Secondary alcohol
- Organoheterocyclic compound
- Oxacycle
- Acetal
- Carboxylic acid derivative
- Hydrocarbon derivative
- Organic oxide
- Organic oxygen compound
- Carbonyl group
- Alcohol
- Organooxygen compound
- Aliphatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aliphatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-056r-7090000000-409b29d29f5d92b036e1 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-06dr-9025400000-db65ccd917ab30b1e85a | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0a6r-0039000000-fd571dbea940514eb044 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a7i-1096000000-473f19a2a3e09916098b | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-014r-3091000000-c7c1e8b2c82cbd9fa2db | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0059-0089000000-7e83fcc43328f0da5170 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-057i-0097000000-41ff44d74a98fc8c733b | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-000l-3190000000-cc31694b1766c38571fa | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-0009000000-860df1894ebe815b95f4 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-2039000000-09a3e28dac6072fa36c4 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-000i-1190000000-ea4455a1fec72fb52745 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-004i-0009000000-0de6306d29da47cbad7b | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-056r-0039000000-e3b3433b12e8c877e6be | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4l-9020000000-72a7050fc022305ce16d | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0036735 |
|---|
| FooDB ID | FDB015672 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | C00011512 |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 475104 |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 11875005 |
|---|
| Kegg Compound ID | C07605 |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|