| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 01:50:33 UTC |
|---|
| Update Date | 2016-11-09 01:19:03 UTC |
|---|
| Accession Number | CHEM030357 |
|---|
| Identification |
|---|
| Common Name | Crispolide |
|---|
| Class | Small Molecule |
|---|
| Description | Crispolide is found in herbs and spices. Crispolide is a constituent of Tanacetum vulgare var. crispum. |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
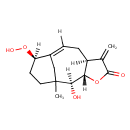
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 1b-Hydroperoxy-5b-hydroxy-4,14-cyclo-9,11-germacradien-12,6a-olide | HMDB |
|
|---|
| Chemical Formula | C15H20O5 |
|---|
| Average Molecular Mass | 280.316 g/mol |
|---|
| Monoisotopic Mass | 280.131 g/mol |
|---|
| CAS Registry Number | 83217-86-1 |
|---|
| IUPAC Name | (2S,3S,7S,11R)-11-hydroperoxy-2-hydroxy-1-methyl-6-methylidene-4-oxatricyclo[8.3.1.0^{3,7}]tetradec-9-en-5-one |
|---|
| Traditional Name | (2S,3S,7S,11R)-11-hydroperoxy-2-hydroxy-1-methyl-6-methylidene-4-oxatricyclo[8.3.1.0^{3,7}]tetradec-9-en-5-one |
|---|
| SMILES | CC12CC[C@@H](OO)C(C1)=CC[C@@H]1[C@H](OC(=O)C1=C)[C@@H]2O |
|---|
| InChI Identifier | InChI=1S/C15H20O5/c1-8-10-4-3-9-7-15(2,6-5-11(9)20-18)13(16)12(10)19-14(8)17/h3,10-13,16,18H,1,4-7H2,2H3/b9-3+/t10-,11+,12-,13-,15?/m0/s1 |
|---|
| InChI Key | JXXWNBNYEWOORY-BASWEGAOSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as germacranolides and derivatives. These are sesquiterpene lactones with a structure based on the germacranolide skeleton, characterized by a gamma lactone fused to a 1,7-dimethylcyclodec-1-ene moiety. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Terpene lactones |
|---|
| Direct Parent | Germacranolides and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Germacranolide
- Sesquiterpenoid
- Gamma butyrolactone
- Tetrahydrofuran
- Enoate ester
- Alpha,beta-unsaturated carboxylic ester
- Carboxylic acid ester
- Lactone
- Hydroperoxide
- Secondary alcohol
- Carboxylic acid derivative
- Peroxol
- Organoheterocyclic compound
- Monocarboxylic acid or derivatives
- Oxacycle
- Alkyl hydroperoxide
- Organooxygen compound
- Organic oxygen compound
- Hydrocarbon derivative
- Organic oxide
- Alcohol
- Carbonyl group
- Aliphatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aliphatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0zg0-1940000000-e030fc44b3940015343a | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-0079-6869000000-00cea548e2553bbd24c5 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-001i-0090000000-ff4cd129993fc0e1342b | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-06sr-0290000000-c7473c8da004393f2239 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0006-3940000000-7f232a2cb895b35704c3 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-0090000000-992a143d3eb7244d48b3 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-01ti-0090000000-2e6408dd809d549b8bf4 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0zfs-9060000000-efe75263a4b311ebb0b5 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0002-0090000000-2cb14853e10b3483a75b | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0002-0090000000-3487fc3ae6103a394c4a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0ab9-1960000000-bb191514460e9a8bec1a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-0090000000-043a95ec3a9c60d0d2b5 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-002b-0090000000-610032dcf8bf88ba5792 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00l7-0190000000-fd68ff99a61f624b358e | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0036695 |
|---|
| FooDB ID | FDB015629 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | C00012420 |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 68894490 |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 131752041 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|