| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 01:48:57 UTC |
|---|
| Update Date | 2016-11-09 01:19:03 UTC |
|---|
| Accession Number | CHEM030316 |
|---|
| Identification |
|---|
| Common Name | Rubinic acid |
|---|
| Class | Small Molecule |
|---|
| Description | Rubinic acid is found in fruits. Rubinic acid is a constituent of Rubus fruticosus (blackberry). |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
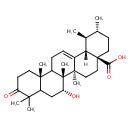
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Rubinate | Generator | | 7a-Hydroxy-3-oxo-12-ursen-28-Oic acid | HMDB | | (1S,2R,4AS,6ar,6BR,7R,12ar,14BS)-7-hydroxy-1,2,6a,6b,9,9,12a-heptamethyl-10-oxo-1,2,3,4,4a,5,6,6a,6b,7,8,8a,9,10,11,12,12a,12b,13,14b-icosahydropicene-4a-carboxylate | Generator |
|
|---|
| Chemical Formula | C30H46O4 |
|---|
| Average Molecular Mass | 470.684 g/mol |
|---|
| Monoisotopic Mass | 470.340 g/mol |
|---|
| CAS Registry Number | 94662-96-1 |
|---|
| IUPAC Name | (1S,2R,4aS,6aR,6bR,7R,12aR,14bS)-7-hydroxy-1,2,6a,6b,9,9,12a-heptamethyl-10-oxo-1,2,3,4,4a,5,6,6a,6b,7,8,8a,9,10,11,12,12a,12b,13,14b-icosahydropicene-4a-carboxylic acid |
|---|
| Traditional Name | (1S,2R,4aS,6aR,6bR,7R,12aR,14bS)-7-hydroxy-1,2,6a,6b,9,9,12a-heptamethyl-10-oxo-1,2,3,4,5,6,7,8,8a,11,12,12b,13,14b-tetradecahydropicene-4a-carboxylic acid |
|---|
| SMILES | [H][C@@]12[C@@H](C)[C@H](C)CC[C@@]1(CC[C@]1(C)C2=CCC2[C@@]3(C)CCC(=O)C(C)(C)C3C[C@@H](O)[C@@]12C)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C30H46O4/c1-17-10-13-30(25(33)34)15-14-28(6)19(24(30)18(17)2)8-9-20-27(5)12-11-22(31)26(3,4)21(27)16-23(32)29(20,28)7/h8,17-18,20-21,23-24,32H,9-16H2,1-7H3,(H,33,34)/t17-,18+,20?,21?,23-,24+,27-,28-,29+,30+/m1/s1 |
|---|
| InChI Key | MFYBXHRQBFYAQZ-MNPGUHGCSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as triterpenoids. These are terpene molecules containing six isoprene units. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Triterpenoids |
|---|
| Direct Parent | Triterpenoids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Triterpenoid
- Cyclic alcohol
- Cyclic ketone
- Secondary alcohol
- Ketone
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Carboxylic acid derivative
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Carbonyl group
- Alcohol
- Aliphatic homopolycyclic compound
|
|---|
| Molecular Framework | Aliphatic homopolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0a5l-0293800000-6832fd6907db8c884fee | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-0f6t-1022192000-b952c2c763cd8e05f46a | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0uk9-0000900000-a61d83ce39c329b7276f | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0pbi-0110900000-5feefdcc676b00b7a9ce | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a6r-5264900000-803bfc6d086d10998bcb | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-014i-0000900000-cde9e843e174885928d8 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-05r0-0000900000-05a07d2d4419b4480621 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0pb9-2000900000-da19bd85b59bbcf26dac | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00di-0000900000-e75dc1fd94a407ce7cce | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0fe0-1441900000-b26be2eb995b6ec7d15d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-05g0-2920000000-1a9f83f4651e3bab1cc9 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-014i-0000900000-d60788820b8857f5a5d5 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-014i-0000900000-af4ea99d509d44286865 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-066r-0000900000-332e470a4471a21931dc | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0036655 |
|---|
| FooDB ID | FDB015581 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | C00057682 |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 35014181 |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 131752024 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|