| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 01:46:49 UTC |
|---|
| Update Date | 2016-11-09 01:19:02 UTC |
|---|
| Accession Number | CHEM030268 |
|---|
| Identification |
|---|
| Common Name | 3'-Hydroxy-T2 Toxin |
|---|
| Class | Small Molecule |
|---|
| Description | Mycotoxin from Fusarium sporotrichiella poae. Metabolite of T2 Toxin. Potential cereal contaminant |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
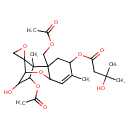
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 3'-Hydroxy T-2 toxin | HMDB | | 3'-Hydroxy T2 toxin | HMDB | | 3'-Hydroxy-T 2 | HMDB | | 3'-Hydroxy-T-2 toxin | HMDB | | 3'-Hydroxy-toxin T-2 | HMDB | | 11'-(Acetyloxy)-2'-[(acetyloxy)methyl]-10'-hydroxy-1',5'-dimethyl-8'-oxaspiro[oxirane-2,12'-tricyclo[7.2.1.0²,⁷]dodecan]-5'-en-4'-yl 3-hydroxy-3-methylbutanoic acid | Generator |
|
|---|
| Chemical Formula | C24H34O10 |
|---|
| Average Molecular Mass | 482.521 g/mol |
|---|
| Monoisotopic Mass | 482.215 g/mol |
|---|
| CAS Registry Number | 84474-35-1 |
|---|
| IUPAC Name | 11'-(acetyloxy)-2'-[(acetyloxy)methyl]-10'-hydroxy-1',5'-dimethyl-8'-oxaspiro[oxirane-2,12'-tricyclo[7.2.1.0²,⁷]dodecan]-5'-en-4'-yl 3-hydroxy-3-methylbutanoate |
|---|
| Traditional Name | 11'-(acetyloxy)-2'-[(acetyloxy)methyl]-10'-hydroxy-1',5'-dimethyl-8'-oxaspiro[oxirane-2,12'-tricyclo[7.2.1.0²,⁷]dodecan]-5'-en-4'-yl 3-hydroxy-3-methylbutanoate |
|---|
| SMILES | CC(=O)OCC12CC(OC(=O)CC(C)(C)O)C(C)=CC1OC1C(O)C(OC(C)=O)C2(C)C11CO1 |
|---|
| InChI Identifier | InChI=1S/C24H34O10/c1-12-7-16-23(10-30-13(2)25,8-15(12)33-17(27)9-21(4,5)29)22(6)19(32-14(3)26)18(28)20(34-16)24(22)11-31-24/h7,15-16,18-20,28-29H,8-11H2,1-6H3 |
|---|
| InChI Key | MLDQZNYFEGLVAS-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as trichothecenes. These are sesquiterpene mycotoxins structurally characterized by the presence of an epoxide ring and a benzopyran derivative with a variant number of hydroxyl, acetyl, or other substituents. The most important structural features causing the biological activities of trichothecenes are the 12,13-epoxy ring, the presence of hydroxyl or acetyl groups at appropriate positions on the trichothecene nucleus and the structure and position of the side-chain. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Sesquiterpenoids |
|---|
| Direct Parent | Trichothecenes |
|---|
| Alternative Parents | |
|---|
| Substituents | - Trichothecene skeleton
- Tricarboxylic acid or derivatives
- Fatty acid ester
- Oxepane
- Oxane
- Fatty acyl
- Cyclic alcohol
- Tertiary alcohol
- Carboxylic acid ester
- Secondary alcohol
- Organoheterocyclic compound
- Carboxylic acid derivative
- Oxacycle
- Dialkyl ether
- Oxirane
- Ether
- Carbonyl group
- Hydrocarbon derivative
- Organic oxygen compound
- Organooxygen compound
- Alcohol
- Organic oxide
- Aliphatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aliphatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0a4i-8147900000-4ad26f37f2a9208363b1 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-03ds-9433643000-3aba3ac90c334dcec824 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0159-1102900000-ca911c50321f80129a88 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0l7i-3316900000-0f288101eaac981f5f94 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0kmi-3409300000-38f1dff96c606e74b946 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-001i-4002900000-1d23e00283846ddf94f2 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0ac0-9205700000-5719c298b9cdd929be7e | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4j-6910000000-412a36007fd68a3f63a7 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-060r-0006900000-d2491c952175e808516f | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0596-2009100000-80c7c53c2ea553be8bd8 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0006-9026300000-11d92a6cb3e782c39ea7 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00di-2003900000-1360593ea64f0492c2bf | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4i-9000000000-470df7e7de367d5d29ed | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4l-9111100000-953867de497d45147c16 | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0036601 |
|---|
| FooDB ID | FDB015516 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | C00012669 |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | Not Available |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 78169621 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|