| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 01:43:04 UTC |
|---|
| Update Date | 2016-11-09 01:19:01 UTC |
|---|
| Accession Number | CHEM030190 |
|---|
| Identification |
|---|
| Common Name | 8-Epixanthatin |
|---|
| Class | Small Molecule |
|---|
| Description | 8-Epixanthatin is found in fats and oils. 8-Epixanthatin is a constituent of Helianthus annuus (sunflower). |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
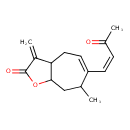
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Xanthatin | MeSH | | 8-Epi-xanthatin | MeSH |
|
|---|
| Chemical Formula | C15H18O3 |
|---|
| Average Molecular Mass | 246.302 g/mol |
|---|
| Monoisotopic Mass | 246.126 g/mol |
|---|
| CAS Registry Number | Not Available |
|---|
| IUPAC Name | 7-methyl-3-methylidene-6-[(1Z)-3-oxobut-1-en-1-yl]-2H,3H,3aH,4H,7H,8H,8aH-cyclohepta[b]furan-2-one |
|---|
| Traditional Name | 7-methyl-3-methylidene-6-[(1Z)-3-oxobut-1-en-1-yl]-3aH,4H,7H,8H,8aH-cyclohepta[b]furan-2-one |
|---|
| SMILES | CC1CC2OC(=O)C(=C)C2CC=C1\C=C/C(C)=O |
|---|
| InChI Identifier | InChI=1S/C15H18O3/c1-9-8-14-13(11(3)15(17)18-14)7-6-12(9)5-4-10(2)16/h4-6,9,13-14H,3,7-8H2,1-2H3/b5-4- |
|---|
| InChI Key | RBRPTFMVULVGIC-PLNGDYQASA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as xanthanolides. These are sesquiterpenoids with a structure based on the xanthanolide skeleton consistsing of a cycloheptane ring usually attached to a four-carbon chain (at carbon 1 ) and a methyl group (at carbon 10), and fused to a five-member lactone ring (sharing carbons 7 and 8). The lactone ring can be conjugated with a methyl or methylene group at position 11. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Terpene lactones |
|---|
| Direct Parent | Xanthanolides |
|---|
| Alternative Parents | |
|---|
| Substituents | - Xanthanolide-skeleton
- Sesquiterpenoid
- Xanthane sesquiterpenoid
- Gamma butyrolactone
- Acryloyl-group
- Enone
- Tetrahydrofuran
- Enoate ester
- Alpha,beta-unsaturated carboxylic ester
- Alpha,beta-unsaturated ketone
- Lactone
- Ketone
- Carboxylic acid ester
- Monocarboxylic acid or derivatives
- Oxacycle
- Carboxylic acid derivative
- Organoheterocyclic compound
- Hydrocarbon derivative
- Organic oxide
- Organooxygen compound
- Organic oxygen compound
- Carbonyl group
- Aliphatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aliphatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-000i-1940000000-838cf5d65c52a850b8f8 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-002b-0390000000-d5170e3e0142f667b18b | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00bj-0940000000-47ca43a9f5ccc49b43b2 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-066r-4900000000-b726394e8925abe2b272 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-0090000000-8bd37f524c1202ced47b | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0f6t-0190000000-d51434285096ea1f55c0 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0f76-6910000000-09e4b8cfd843959f3fce | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-0090000000-3af6d9b55981b63a31cc | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004j-3490000000-a8a35b318c968e285bf5 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0100-3900000000-cb093a29a945ca6d323d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000b-0590000000-c062b73cb301a428b090 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0fb9-0960000000-4d3c8bee9d38942b676f | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-002f-5910000000-2c404c8d13a163638f06 | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0036473 |
|---|
| FooDB ID | FDB015367 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | C00000497 |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 35014156 |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 131751996 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|