| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 01:38:17 UTC |
|---|
| Update Date | 2016-11-09 01:19:00 UTC |
|---|
| Accession Number | CHEM030075 |
|---|
| Identification |
|---|
| Common Name | Ethyl 7-epi-12-hydroxyjasmonate glucoside |
|---|
| Class | Small Molecule |
|---|
| Description | Ethyl 7-epi-12-hydroxyjasmonate glucoside is found in herbs and spices. Ethyl 7-epi-12-hydroxyjasmonate glucoside is a constituent of sage (Salvia officinalis) |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
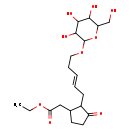
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Ethyl 7-epi-12-hydroxyjasmonic acid glucoside | Generator | | Ethyl 2-{3-oxo-2-[(2E)-5-{[3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy}pent-2-en-1-yl]cyclopentyl}acetic acid | Generator |
|
|---|
| Chemical Formula | C20H32O9 |
|---|
| Average Molecular Mass | 416.463 g/mol |
|---|
| Monoisotopic Mass | 416.205 g/mol |
|---|
| CAS Registry Number | Not Available |
|---|
| IUPAC Name | ethyl 2-{3-oxo-2-[(2E)-5-{[3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy}pent-2-en-1-yl]cyclopentyl}acetate |
|---|
| Traditional Name | ethyl 2-{3-oxo-2-[(2E)-5-{[3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy}pent-2-en-1-yl]cyclopentyl}acetate |
|---|
| SMILES | CCOC(=O)CC1CCC(=O)C1C\C=C\CCOC1OC(CO)C(O)C(O)C1O |
|---|
| InChI Identifier | InChI=1S/C20H32O9/c1-2-27-16(23)10-12-7-8-14(22)13(12)6-4-3-5-9-28-20-19(26)18(25)17(24)15(11-21)29-20/h3-4,12-13,15,17-21,24-26H,2,5-11H2,1H3/b4-3+ |
|---|
| InChI Key | DALGUVBWVCFIPV-ONEGZZNKSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as fatty acyl glycosides of mono- and disaccharides. Fatty acyl glycosides of mono- and disaccharides are compounds composed of a mono- or disaccharide moiety linked to one hydroxyl group of a fatty alcohol or of a phosphorylated alcohol (phosphoprenols), a hydroxy fatty acid or to one carboxyl group of a fatty acid (ester linkage) or to an amino alcohol. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Fatty Acyls |
|---|
| Sub Class | Fatty acyl glycosides |
|---|
| Direct Parent | Fatty acyl glycosides of mono- and disaccharides |
|---|
| Alternative Parents | |
|---|
| Substituents | - Fatty acyl glycoside of mono- or disaccharide
- Jasmonic acid
- Alkyl glycoside
- Hexose monosaccharide
- Glycosyl compound
- O-glycosyl compound
- Monosaccharide
- Oxane
- Carboxylic acid ester
- Ketone
- Cyclic ketone
- Secondary alcohol
- Monocarboxylic acid or derivatives
- Organoheterocyclic compound
- Oxacycle
- Acetal
- Carboxylic acid derivative
- Polyol
- Alcohol
- Hydrocarbon derivative
- Organic oxide
- Carbonyl group
- Organic oxygen compound
- Organooxygen compound
- Primary alcohol
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-05g1-7329000000-145c174f02833d43c319 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (3 TMS) - 70eV, Positive | splash10-014i-4295057000-0cdb1946634a77dfbfb8 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014j-0129300000-996eb878e66c13c155f5 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-06rb-5975000000-5ef470e5915151216c8c | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-01qa-9813000000-bb0e4a70d6111e054b87 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-014i-5578900000-4dc3622060f41723d039 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0401-6924000000-fce8220e95f09c6c8f97 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0k97-9230000000-26ae5eecb25d3ccb33d3 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-014i-0101900000-5959bb13cedd2e7cb193 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-05ox-4109000000-319b1a7e00246d4965bf | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0036-5946000000-5ce0ad0c483a2f3ded08 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0002-0109100000-72304a5f3bb522514fb4 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0fxw-3339000000-9da84f542048204c9f4c | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4l-9023000000-e41798378b9a757fb3f6 | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0036340 |
|---|
| FooDB ID | FDB015211 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 74886455 |
|---|
| ChEBI ID | 168758 |
|---|
| PubChem Compound ID | 131751966 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|