| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 01:37:05 UTC |
|---|
| Update Date | 2016-11-09 01:19:00 UTC |
|---|
| Accession Number | CHEM030041 |
|---|
| Identification |
|---|
| Common Name | Ganoderic acid delta |
|---|
| Class | Small Molecule |
|---|
| Description | Constituent of Ganoderma lucidum (reishi). Ganoderic acid delta is found in mushrooms. |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
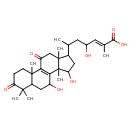
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Ganoderate delta | Generator | | Ganoderate δ | Generator | | Ganoderic acid δ | Generator | | 7beta,12alpha-Dihydroxy-3,11,23-tetraoxo-5alpha-lanost-8-en-26-Oic acid | HMDB | | Ganoderic acid D | HMDB | | (2E)-6-{9,12-dihydroxy-2,6,6,11,15-pentamethyl-5,17-dioxotetracyclo[8.7.0.0²,⁷.0¹¹,¹⁵]heptadec-1(10)-en-14-yl}-4-hydroxy-2-methylhept-2-enoate | Generator |
|
|---|
| Chemical Formula | C30H44O7 |
|---|
| Average Molecular Mass | 516.666 g/mol |
|---|
| Monoisotopic Mass | 516.309 g/mol |
|---|
| CAS Registry Number | 294674-02-5 |
|---|
| IUPAC Name | (2E)-6-{9,12-dihydroxy-2,6,6,11,15-pentamethyl-5,17-dioxotetracyclo[8.7.0.0²,⁷.0¹¹,¹⁵]heptadec-1(10)-en-14-yl}-4-hydroxy-2-methylhept-2-enoic acid |
|---|
| Traditional Name | (2E)-6-{9,12-dihydroxy-2,6,6,11,15-pentamethyl-5,17-dioxotetracyclo[8.7.0.0²,⁷.0¹¹,¹⁵]heptadec-1(10)-en-14-yl}-4-hydroxy-2-methylhept-2-enoic acid |
|---|
| SMILES | CC(CC(O)\C=C(/C)C(O)=O)C1CC(O)C2(C)C3=C(C(=O)CC12C)C1(C)CCC(=O)C(C)(C)C1CC3O |
|---|
| InChI Identifier | InChI=1S/C30H44O7/c1-15(10-17(31)11-16(2)26(36)37)18-12-23(35)30(7)25-19(32)13-21-27(3,4)22(34)8-9-28(21,5)24(25)20(33)14-29(18,30)6/h11,15,17-19,21,23,31-32,35H,8-10,12-14H2,1-7H3,(H,36,37)/b16-11+ |
|---|
| InChI Key | JTBDTJVGIGEPFI-LFIBNONCSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as triterpenoids. These are terpene molecules containing six isoprene units. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Triterpenoids |
|---|
| Direct Parent | Triterpenoids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Triterpenoid
- Trihydroxy bile acid, alcohol, or derivatives
- Hydroxy bile acid, alcohol, or derivatives
- 23-hydroxysteroid
- Bile acid, alcohol, or derivatives
- Steroid acid
- 3-oxosteroid
- 15-hydroxysteroid
- Hydroxysteroid
- 11-oxosteroid
- Oxosteroid
- 7-hydroxysteroid
- Steroid
- Medium-chain hydroxy acid
- Medium-chain fatty acid
- Hydroxy fatty acid
- Cyclohexenone
- Fatty acid
- Unsaturated fatty acid
- Fatty acyl
- Cyclic alcohol
- Ketone
- Secondary alcohol
- Cyclic ketone
- Carboxylic acid
- Carboxylic acid derivative
- Monocarboxylic acid or derivatives
- Carbonyl group
- Alcohol
- Organic oxide
- Hydrocarbon derivative
- Organic oxygen compound
- Organooxygen compound
- Aliphatic homopolycyclic compound
|
|---|
| Molecular Framework | Aliphatic homopolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-00dj-0125920000-56ce74044f43756df6e9 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-0002-1210219000-a9d3ca84a6f1358ecb2b | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-001j-0000910000-5a4da39b5a6a85aa5322 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0fh9-2000900000-9afa8f3947a679d9bf6e | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0ugi-1102900000-88e68243a71603ee2cef | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-014j-1000980000-397a963e0a457b8680d1 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0v4j-2000910000-fa008150f11068dcb4be | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0avi-9011700000-6bb6303e15316e65a6c7 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00lj-0408920000-1044b2df28df5310eff1 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00kr-4809610000-9278a449b28d8b75a82f | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000i-5209200000-a2172f397711a0c820b5 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-014i-0000390000-7725b4dc90e6812daa49 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0uk9-1001910000-9ed4496d2e17787c87da | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0udr-2002900000-068bcdca76aa826590c0 | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0036306 |
|---|
| FooDB ID | FDB015174 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | Not Available |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 131751950 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|