| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 01:34:42 UTC |
|---|
| Update Date | 2016-11-09 01:18:59 UTC |
|---|
| Accession Number | CHEM029986 |
|---|
| Identification |
|---|
| Common Name | Maleic acid homopolymer |
|---|
| Class | Small Molecule |
|---|
| Description | A dicarboxylic acid that is maleic acid in which each of the hydrogens that is attached to a carbon atom is substituted by a methyl group. |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
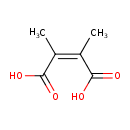
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 2,3-Dimethylmaleic acid | ChEBI | | alpha,beta-Dimethylmaleic acid | ChEBI | | Dimethylmaleic acid | Kegg | | 2,3-Dimethylmaleate | Generator | | a,b-Dimethylmaleate | Generator | | a,b-Dimethylmaleic acid | Generator | | alpha,beta-Dimethylmaleate | Generator | | Α,β-dimethylmaleate | Generator | | Α,β-dimethylmaleic acid | Generator | | Dimethylmaleate | Generator | | Maleate homopolymer | Generator | | meso-Tetra(4-sulfonatophenyl)porphine, tetrasodium salt | HMDB | | meso-Tetra(P-sulfonatophenyl)porphine | HMDB | | meso-Tetra-(4-sulfonatophenyl)porphine | HMDB | | meso-Tetra-(4-sulphonatophenyl) porphine | HMDB | | meso-Tetrakis(P-sulfophenyl)porphine tetrasodium salt | HMDB | | meso-Tetraphenylporphine-4,4',4",4"'-tetrasulfonic acid | HMDB | | SFP | HMDB | | Tetraphenylporphine sulfonate | HMDB | | Tetrasodium meso-tetra(P-sulfonatophenyl)porphine | HMDB | | TPPS (Photoreactant) | HMDB | | TPPS4 | HMDB |
|
|---|
| Chemical Formula | C6H8O4 |
|---|
| Average Molecular Mass | 144.125 g/mol |
|---|
| Monoisotopic Mass | 144.042 g/mol |
|---|
| CAS Registry Number | 488-21-1 |
|---|
| IUPAC Name | (2Z)-dimethylbut-2-enedioic acid |
|---|
| Traditional Name | dimethylmaleic acid |
|---|
| SMILES | C\C(=C(/C)C(O)=O)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C6H8O4/c1-3(5(7)8)4(2)6(9)10/h1-2H3,(H,7,8)(H,9,10)/b4-3- |
|---|
| InChI Key | CGBYBGVMDAPUIH-ARJAWSKDSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as methyl-branched fatty acids. These are fatty acids with an acyl chain that has a methyl branch. Usually, they are saturated and contain only one or more methyl group. However, branches other than methyl may be present. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Fatty Acyls |
|---|
| Sub Class | Fatty acids and conjugates |
|---|
| Direct Parent | Methyl-branched fatty acids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Methyl-branched fatty acid
- Unsaturated fatty acid
- Dicarboxylic acid or derivatives
- Carboxylic acid
- Carboxylic acid derivative
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Carbonyl group
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0f97-9300000000-ca4db8fbae55af356262 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-00di-9430000000-cc5c14a34aebae0b44cd | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0002-5900000000-47d64c9efeb2425c6a25 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-05i1-9100000000-3907b4f957e123f00f06 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0udi-9000000000-fa4ea12762e730f639c5 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0006-4900000000-030a507d026921a19b06 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0007-9400000000-8fd12833fc2b1eb092b6 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0pi0-9000000000-042313ce67784ad19929 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-006w-9400000000-91b99e534309dd012bd1 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-05fs-9000000000-08c30e63eaf1e8bee433 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-9000000000-69ebf0092adf747aaffc | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-003s-9300000000-f5922c36d63b69918426 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0pc0-9000000000-6f11bc7a996759e3b0b7 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0zfr-9000000000-dc48b96f3be320b2477a | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0036232 |
|---|
| FooDB ID | FDB015090 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | DIMETHYLMAL-CPD |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 2297318 |
|---|
| ChEBI ID | 23812 |
|---|
| PubChem Compound ID | 3032309 |
|---|
| Kegg Compound ID | C00922 |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|