| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 01:33:13 UTC |
|---|
| Update Date | 2016-11-09 01:18:59 UTC |
|---|
| Accession Number | CHEM029967 |
|---|
| Identification |
|---|
| Common Name | 4,11,13,15-Tetrahydroridentin B |
|---|
| Class | Small Molecule |
|---|
| Description | 4,11,13,15-Tetrahydroridentin B is found in alcoholic beverages. 4,11,13,15-Tetrahydroridentin B is a constituent of Taraxacum officinale (dandelion) |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
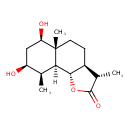
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 4alpha(15), 11beta(13)-Tetrahydroridentin b | HMDB |
|
|---|
| Chemical Formula | C15H24O4 |
|---|
| Average Molecular Mass | 268.349 g/mol |
|---|
| Monoisotopic Mass | 268.167 g/mol |
|---|
| CAS Registry Number | 75991-58-1 |
|---|
| IUPAC Name | (3S,3aS,5aR,6R,8S,9R,9aS,9bS)-6,8-dihydroxy-3,5a,9-trimethyl-dodecahydronaphtho[1,2-b]furan-2-one |
|---|
| Traditional Name | (3S,3aS,5aR,6R,8S,9R,9aS,9bS)-6,8-dihydroxy-3,5a,9-trimethyl-decahydronaphtho[1,2-b]furan-2-one |
|---|
| SMILES | [H][C@@]12[C@H]3OC(=O)[C@@H](C)[C@@H]3CC[C@@]1(C)[C@H](O)C[C@H](O)[C@@H]2C |
|---|
| InChI Identifier | InChI=1S/C15H24O4/c1-7-9-4-5-15(3)11(17)6-10(16)8(2)12(15)13(9)19-14(7)18/h7-13,16-17H,4-6H2,1-3H3/t7-,8-,9-,10-,11+,12+,13-,15-/m0/s1 |
|---|
| InChI Key | GAPZIAIPNAGPQZ-UIUPBXLSSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as eudesmanolides, secoeudesmanolides, and derivatives. These are terpenoids with a structure based on the eudesmanolide (a 3,5a,9-trimethyl-naphtho[1,2-b]furan-2-one derivative) or secoeudesmanolide (a 3,6-dimethyl-5-(pentan-2-yl)-1-benzofuran-2-one derivative) skeleton. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Terpene lactones |
|---|
| Direct Parent | Eudesmanolides, secoeudesmanolides, and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Eudesmanolide
- Sesquiterpenoid
- Naphthofuran
- Gamma butyrolactone
- Cyclic alcohol
- Tetrahydrofuran
- Secondary alcohol
- Lactone
- Carboxylic acid ester
- Oxacycle
- Monocarboxylic acid or derivatives
- Carboxylic acid derivative
- Organoheterocyclic compound
- Organooxygen compound
- Hydrocarbon derivative
- Organic oxide
- Organic oxygen compound
- Alcohol
- Carbonyl group
- Aliphatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aliphatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0fk9-0890000000-f088b10a81ba7c1eaa7a | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-0092-2139000000-d03f1ab72fc5e1cd7f39 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0uxr-0190000000-5109f0fa30402e9a8635 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0v0r-0960000000-4f0c1b31671a406e965d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00kr-5910000000-6ef176ef49116a20ae8c | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-014i-0090000000-e167a33bb536de00ee04 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00kb-0190000000-c1bccbcae119e234dc48 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0v4i-2930000000-1b40b8bc6c01606eb86c | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014i-0390000000-8200856d8403c07384f5 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0gbd-0690000000-f2e0d60cd18baa32abd8 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000j-2920000000-6c86463a18d12d856599 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-014i-0090000000-9699a607ea019e68b5c3 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-014i-0090000000-a398947045938d7308a3 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-066r-3590000000-72e3a90d5db71e242b49 | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0036150 |
|---|
| FooDB ID | FDB015001 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | C00058192 |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 30777158 |
|---|
| ChEBI ID | 169733 |
|---|
| PubChem Compound ID | 15540432 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | | 1. Simons K, Toomre D: Lipid rafts and signal transduction. Nat Rev Mol Cell Biol. 2000 Oct;1(1):31-9. | | 2. Watson AD: Thematic review series: systems biology approaches to metabolic and cardiovascular disorders. Lipidomics: a global approach to lipid analysis in biological systems. J Lipid Res. 2006 Oct;47(10):2101-11. Epub 2006 Aug 10. | | 3. Sethi JK, Vidal-Puig AJ: Thematic review series: adipocyte biology. Adipose tissue function and plasticity orchestrate nutritional adaptation. J Lipid Res. 2007 Jun;48(6):1253-62. Epub 2007 Mar 20. | | 4. Lingwood D, Simons K: Lipid rafts as a membrane-organizing principle. Science. 2010 Jan 1;327(5961):46-50. doi: 10.1126/science.1174621. | | 5. Yannai, Shmuel. (2004) Dictionary of food compounds with CD-ROM: Additives, flavors, and ingredients. Boca Raton: Chapman & Hall/CRC. | | 6. Zielinska, K., Kisiel, W., Sesquiterpenoids from roots of Taraxacum laevigatum and Taraxacum disseminatum, Phytochemistry 54 (2000) 791-794 [Structure] | | 7. The lipid handbook with CD-ROM |
|
|---|