| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 01:33:09 UTC |
|---|
| Update Date | 2016-11-09 01:18:59 UTC |
|---|
| Accession Number | CHEM029965 |
|---|
| Identification |
|---|
| Common Name | Furanojaponin |
|---|
| Class | Small Molecule |
|---|
| Description | Furanojaponin is found in giant butterbur. Furanojaponin is a constituent of Petasites japonicus (sweet coltsfoot) |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
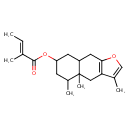
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 3,4a,5-Trimethyl-4H,4ah,5H,6H,7H,8H,8ah,9H-naphtho[2,3-b]furan-7-yl (2E)-2-methylbut-2-enoic acid | HMDB |
|
|---|
| Chemical Formula | C20H28O3 |
|---|
| Average Molecular Mass | 316.435 g/mol |
|---|
| Monoisotopic Mass | 316.204 g/mol |
|---|
| CAS Registry Number | 34335-98-3 |
|---|
| IUPAC Name | 3,4a,5-trimethyl-4H,4aH,5H,6H,7H,8H,8aH,9H-naphtho[2,3-b]furan-7-yl (2E)-2-methylbut-2-enoate |
|---|
| Traditional Name | 3,4a,5-trimethyl-4H,5H,6H,7H,8H,8aH,9H-naphtho[2,3-b]furan-7-yl (2E)-2-methylbut-2-enoate |
|---|
| SMILES | C\C=C(/C)C(=O)OC1CC(C)C2(C)CC3=C(CC2C1)OC=C3C |
|---|
| InChI Identifier | InChI=1S/C20H28O3/c1-6-12(2)19(21)23-16-7-14(4)20(5)10-17-13(3)11-22-18(17)9-15(20)8-16/h6,11,14-16H,7-10H2,1-5H3/b12-6+ |
|---|
| InChI Key | DDJITDJHDCLHOK-WUXMJOGZSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as eremophilane, 8,9-secoeremophilane and furoeremophilane sesquiterpenoids. These are sesquiterpenoids with a structure based either on the eremophilane skeleton, its 8,9-seco derivative, or the furoeremophilane skeleton. Eremophilanes have been shown to be derived from eudesmanes by migration of the methyl group at C-10 to C-5. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Sesquiterpenoids |
|---|
| Direct Parent | Eremophilane, 8,9-secoeremophilane and furoeremophilane sesquiterpenoids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Furoeremophilane sesquiterpenoid
- Naphthofuran
- Benzofuran
- Fatty acid ester
- Fatty acyl
- Furan
- Enoate ester
- Alpha,beta-unsaturated carboxylic ester
- Heteroaromatic compound
- Carboxylic acid ester
- Monocarboxylic acid or derivatives
- Carboxylic acid derivative
- Organoheterocyclic compound
- Oxacycle
- Organic oxygen compound
- Organooxygen compound
- Hydrocarbon derivative
- Organic oxide
- Carbonyl group
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-053i-4911000000-1e6c57e1825ab304a4b1 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014i-4159000000-64d49d3ff0bc594942cf | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-067i-9351000000-9b961376b72ede30801f | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0f89-9200000000-4f6fe44eefc351f2fbb9 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-014i-1029000000-24ba7215de5ef8968ad9 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0159-9087000000-b0ea859edd98a719908c | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0zgj-8390000000-f132c3a6942bd8aa6b7f | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00kr-0091000000-45751ff543da86e979a2 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-02t9-2490000000-19afb4a241fb41157cc3 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0cdj-4940000000-b076d060fc260bf4b69a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-001j-8092000000-f009227d523375e0c67f | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0002-9000000000-86a2ef610e54952992c9 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0gb9-5090000000-5722a034bcb9587c1ed6 | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0036147 |
|---|
| FooDB ID | FDB014998 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | C00017364 |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 35014103 |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 131751918 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|