| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 01:32:31 UTC |
|---|
| Update Date | 2016-11-09 01:18:59 UTC |
|---|
| Accession Number | CHEM029952 |
|---|
| Identification |
|---|
| Common Name | Demethyloleuropein |
|---|
| Class | Small Molecule |
|---|
| Description | Demethyloleuropein is found in fruits. Demethyloleuropein is present in olive fruits and the bark of Syringa vulgaris. Indicator of maturity in olives which increases as the fruit ripen |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
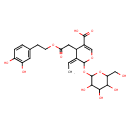
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 4-{2-[2-(3,4-dihydroxyphenyl)ethoxy]-2-oxoethyl}-3-ethylidene-2-{[3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy}-3,4-dihydro-2H-pyran-5-carboxylate | HMDB | | Demethyloleuropein | MeSH |
|
|---|
| Chemical Formula | C24H30O13 |
|---|
| Average Molecular Mass | 526.487 g/mol |
|---|
| Monoisotopic Mass | 526.169 g/mol |
|---|
| CAS Registry Number | 52077-55-1 |
|---|
| IUPAC Name | (3Z)-4-{2-[2-(3,4-dihydroxyphenyl)ethoxy]-2-oxoethyl}-3-ethylidene-2-{[3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy}-3,4-dihydro-2H-pyran-5-carboxylic acid |
|---|
| Traditional Name | (5Z)-4-{2-[2-(3,4-dihydroxyphenyl)ethoxy]-2-oxoethyl}-5-ethylidene-6-{[3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy}-4,6-dihydropyran-3-carboxylic acid |
|---|
| SMILES | [H][C@]1(CC(=O)OCCC2=CC(O)=C(O)C=C2)\C(=C/C)[C@H](O[C@@H]2O[C@H](CO)[C@@H](O)[C@H](O)[C@H]2O)OC=C1C(O)=O |
|---|
| InChI Identifier | InChI=1S/C24H30O13/c1-2-12-13(8-18(28)34-6-5-11-3-4-15(26)16(27)7-11)14(22(32)33)10-35-23(12)37-24-21(31)20(30)19(29)17(9-25)36-24/h2-4,7,10,13,17,19-21,23-27,29-31H,5-6,8-9H2,1H3,(H,32,33)/b12-2+/t13-,17+,19+,20-,21+,23-,24-/m0/s1 |
|---|
| InChI Key | HKVGJQVJNQRJPO-VTDDDATNSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as terpene glycosides. These are prenol lipids containing a carbohydrate moiety glycosidically bound to a terpene backbone. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Terpene glycosides |
|---|
| Direct Parent | Terpene glycosides |
|---|
| Alternative Parents | |
|---|
| Substituents | - Terpene glycoside
- O-glycosyl compound
- Secoiridoid-skeleton
- Glycosyl compound
- Monoterpenoid
- Aromatic monoterpenoid
- Monocyclic monoterpenoid
- Tyrosol derivative
- Catechol
- Sugar acid
- Phenol
- 1-hydroxy-2-unsubstituted benzenoid
- 1-hydroxy-4-unsubstituted benzenoid
- Dicarboxylic acid or derivatives
- Monosaccharide
- Oxane
- Monocyclic benzene moiety
- Benzenoid
- Vinylogous ester
- Secondary alcohol
- Carboxylic acid ester
- Acetal
- Carboxylic acid derivative
- Carboxylic acid
- Oxacycle
- Organoheterocyclic compound
- Polyol
- Hydrocarbon derivative
- Alcohol
- Organic oxygen compound
- Organic oxide
- Primary alcohol
- Carbonyl group
- Organooxygen compound
- Aromatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aromatic heteromonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0ab9-6915520000-ef13bacc01c1c99ec6b0 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-0abj-4932347000-e634412f037ae938d94f | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03fr-0419240000-6ecbd1f9d6f1306267b1 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-08mi-1519210000-0dbd9718a68813e825a0 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-01ox-7968000000-fe227c88df131e39bfce | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-014i-1149000000-3ceba264039055cc33e5 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0zfr-4918200000-b648fd34ef380c834c64 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0600-7927100000-e9495b50b17831be7e9e | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-07xs-0749150000-cfa4191366e92b2029d3 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-029j-0916000000-35fb3716330b63579cc1 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0019-5921000000-7797d26b53c647944e30 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-004j-0509020000-580eda502dcfc3be02ca | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000b-0906000000-92566e4c9cf005cd05fe | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00ri-5901000000-7614cd3f7f9c2de5cf06 | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0036121 |
|---|
| FooDB ID | FDB014969 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | C00055262 |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | Not Available |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 89887620 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|