| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 01:30:46 UTC |
|---|
| Update Date | 2016-11-09 01:18:58 UTC |
|---|
| Accession Number | CHEM029918 |
|---|
| Identification |
|---|
| Common Name | 27-Hydroxyisomangiferolic acid |
|---|
| Class | Small Molecule |
|---|
| Description | 27-Hydroxymangiferolic acid is found in fruits. 27-Hydroxymangiferolic acid is a constituent of the stem bark of Mangifera indica (mango). |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
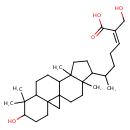
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 27-Hydroxyisomangiferolate | Generator | | Coenzyme Q6 | HMDB | | Coenzyme QQ6 | HMDB | | Coenzyme-Q6 | HMDB | | CoQ-6 | HMDB | | CoQ6 | HMDB | | Ubiquinone 30 | HMDB | | Ubiquinone Q6 | HMDB | | Ubiquinone(6) | HMDB | | UBIQUINONE-6 | HMDB | | (2Z)-6-{6-hydroxy-7,7,12,16-tetramethylpentacyclo[9.7.0.0¹,³.0³,⁸.0¹²,¹⁶]octadecan-15-yl}-2-(hydroxymethyl)hept-2-enoate | HMDB | | 27-Hydroxyisomangiferolic acid | MeSH |
|
|---|
| Chemical Formula | C30H48O4 |
|---|
| Average Molecular Mass | 472.700 g/mol |
|---|
| Monoisotopic Mass | 472.355 g/mol |
|---|
| CAS Registry Number | 123563-64-4 |
|---|
| IUPAC Name | (2Z)-6-{6-hydroxy-7,7,12,16-tetramethylpentacyclo[9.7.0.0¹,³.0³,⁸.0¹²,¹⁶]octadecan-15-yl}-2-(hydroxymethyl)hept-2-enoic acid |
|---|
| Traditional Name | (2Z)-6-{6-hydroxy-7,7,12,16-tetramethylpentacyclo[9.7.0.0¹,³.0³,⁸.0¹²,¹⁶]octadecan-15-yl}-2-(hydroxymethyl)hept-2-enoic acid |
|---|
| SMILES | CC(CC\C=C(\CO)C(O)=O)C1CCC2(C)C3CCC4C5(CC35CCC12C)CCC(O)C4(C)C |
|---|
| InChI Identifier | InChI=1S/C30H48O4/c1-19(7-6-8-20(17-31)25(33)34)21-11-13-28(5)23-10-9-22-26(2,3)24(32)12-14-29(22)18-30(23,29)16-15-27(21,28)4/h8,19,21-24,31-32H,6-7,9-18H2,1-5H3,(H,33,34)/b20-8- |
|---|
| InChI Key | YWPLTMNXKKXXII-ZBKNUEDVSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as cycloartanols and derivatives. These are steroids containing a cycloartanol moiety. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Steroids and steroid derivatives |
|---|
| Sub Class | Cycloartanols and derivatives |
|---|
| Direct Parent | Cycloartanols and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Cycloartanol-skeleton
- 9b,19-cyclo-lanostane-skeleton
- Cycloartane-skeleton
- Triterpenoid
- 26-hydroxysteroid
- Dihydroxy bile acid, alcohol, or derivatives
- Hydroxy bile acid, alcohol, or derivatives
- Bile acid, alcohol, or derivatives
- Steroid acid
- 3-hydroxysteroid
- Hydroxysteroid
- Medium-chain fatty acid
- Beta-hydroxy acid
- Hydroxy fatty acid
- Fatty acyl
- Fatty acid
- Unsaturated fatty acid
- Hydroxy acid
- Cyclic alcohol
- Secondary alcohol
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Carboxylic acid derivative
- Alcohol
- Primary alcohol
- Hydrocarbon derivative
- Carbonyl group
- Organic oxide
- Organic oxygen compound
- Organooxygen compound
- Aliphatic homopolycyclic compound
|
|---|
| Molecular Framework | Aliphatic homopolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-052f-0012900000-a791e328509c821f1256 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (3 TMS) - 70eV, Positive | splash10-00di-1011119000-9758143278dc514e911c | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0a4i-0000900000-8c24f11b6046cafaf49d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a4r-0005900000-b5227b2307ffc877aa84 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4i-2019700000-052b5af0c615f822a34e | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00di-0000900000-db24a78feb89690fb106 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-05i3-0001900000-9c6e897b687f237c791b | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0007-8005900000-d0a06cd832d6b66bdd8c | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-1902400000-ca6ee32eb9dfe748429a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03e9-3729600000-3e69f2db5ae575b9e72f | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03l0-9148100000-a723cd69798f14ed256d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00di-0000900000-c28ab658f937840b7228 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4i-0003900000-32ce53ad6db413578d06 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0002-0109500000-c9ac72c37950e265f822 | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0036064 |
|---|
| FooDB ID | FDB014890 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | C00056698 |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 72390814 |
|---|
| ChEBI ID | 52971 |
|---|
| PubChem Compound ID | 5283544 |
|---|
| Kegg Compound ID | C17568 |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|