| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 01:22:04 UTC |
|---|
| Update Date | 2016-11-09 01:18:56 UTC |
|---|
| Accession Number | CHEM029723 |
|---|
| Identification |
|---|
| Common Name | Scirpene-3,4,15-triol |
|---|
| Class | Small Molecule |
|---|
| Description | Production by Fusarium roseum, Fusarium equiseti and Fusarium sporotrichiella. Mycotoxin |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
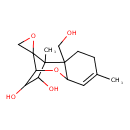
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 12,13-Epoxy-trichothec-9-ene-3alpha,4beta,15-triol | HMDB | | Anguidine deriv scirpentriol | HMDB | | Scirpene-3,4,15-triol | HMDB | | Scirpenetriol | HMDB | | Scirpenol | HMDB | | Scirpentriol | HMDB | | T-2 Triol | MeSH | | 3,4,15-Trihydroxy-8-(3-methylbutyryloxy)-1,2,3-epoxytrichothec--9-ene | MeSH |
|
|---|
| Chemical Formula | C15H22O5 |
|---|
| Average Molecular Mass | 282.332 g/mol |
|---|
| Monoisotopic Mass | 282.147 g/mol |
|---|
| CAS Registry Number | 2270-41-9 |
|---|
| IUPAC Name | 2'-(hydroxymethyl)-1',5'-dimethyl-8'-oxaspiro[oxirane-2,12'-tricyclo[7.2.1.0²,⁷]dodecan]-5'-ene-10',11'-diol |
|---|
| Traditional Name | 2'-(hydroxymethyl)-1',5'-dimethyl-8'-oxaspiro[oxirane-2,12'-tricyclo[7.2.1.0²,⁷]dodecan]-5'-ene-10',11'-diol |
|---|
| SMILES | CC1=CC2OC3C(O)C(O)C(C)(C33CO3)C2(CO)CC1 |
|---|
| InChI Identifier | InChI=1S/C15H22O5/c1-8-3-4-14(6-16)9(5-8)20-12-10(17)11(18)13(14,2)15(12)7-19-15/h5,9-12,16-18H,3-4,6-7H2,1-2H3 |
|---|
| InChI Key | PXEBOIUZEXXBGH-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as trichothecenes. These are sesquiterpene mycotoxins structurally characterized by the presence of an epoxide ring and a benzopyran derivative with a variant number of hydroxyl, acetyl, or other substituents. The most important structural features causing the biological activities of trichothecenes are the 12,13-epoxy ring, the presence of hydroxyl or acetyl groups at appropriate positions on the trichothecene nucleus and the structure and position of the side-chain. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Sesquiterpenoids |
|---|
| Direct Parent | Trichothecenes |
|---|
| Alternative Parents | Not Available |
|---|
| Substituents | Not Available |
|---|
| Molecular Framework | Aliphatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0fk9-3980000000-70e0c82b919ac960d454 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (3 TMS) - 70eV, Positive | splash10-000t-7964800000-3fe4b14d05f42aabb777 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0159-0090000000-464ef52cd83df1b85f0b | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-066s-1590000000-80f55d84841d3b245048 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0gba-9640000000-31027155a4571b4c96c1 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-001i-0090000000-dafc5573bf4d9a60e7dd | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0gx0-0390000000-441c56c96bb039aec8cf | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4j-6900000000-783e38eac8480236c061 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-001i-0090000000-4206ab54dd729eeefe27 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-001i-0190000000-a703d689dc48fee95783 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0uec-1390000000-eef9ee988a3cf07e6528 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-001i-0090000000-0d73455ff384d5026de7 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00lr-0190000000-7cb1ce92533c3aa5266c | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0zgi-4090000000-4789e9a33c6444e84c05 | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0035846 |
|---|
| FooDB ID | FDB014624 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | C00012642 |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 2807424 |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 3570295 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|