| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 01:18:47 UTC |
|---|
| Update Date | 2016-11-09 01:18:55 UTC |
|---|
| Accession Number | CHEM029652 |
|---|
| Identification |
|---|
| Common Name | Brein |
|---|
| Class | Small Molecule |
|---|
| Description | Constituent of Manila elemi resin and oil (Canarium communis), Elemi oil has a citrus-like smell, a bit spicy and is pale in color. In the Philippines the Elemi tree is known locally as 'Pili". The main chemical components of elemi oil are terpineol, elemicine, elemol, dipentene, phellandrene and limonene. It is also found in Baccharis rhomboidalis (a spice shrub primarily found in Chile), Euphorbia species (also commonly referred to as "Spurges"), Farfugium species and others. |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
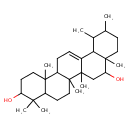
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 12-Ursene-3b,16b-diol | HMDB |
|
|---|
| Chemical Formula | C30H50O2 |
|---|
| Average Molecular Mass | 442.717 g/mol |
|---|
| Monoisotopic Mass | 442.381 g/mol |
|---|
| CAS Registry Number | 31575-82-3 |
|---|
| IUPAC Name | 4,4,6a,6b,8a,11,12,14b-octamethyl-1,2,3,4,4a,5,6,6a,6b,7,8,8a,9,10,11,12,12a,14,14a,14b-icosahydropicene-3,8-diol |
|---|
| Traditional Name | 4,4,6a,6b,8a,11,12,14b-octamethyl-2,3,4a,5,6,7,8,9,10,11,12,12a,14,14a-tetradecahydro-1H-picene-3,8-diol |
|---|
| SMILES | CC1CCC2(C)C(O)CC3(C)C(=CCC4C5(C)CCC(O)C(C)(C)C5CCC34C)C2C1C |
|---|
| InChI Identifier | InChI=1S/C30H50O2/c1-18-11-14-28(6)24(32)17-30(8)20(25(28)19(18)2)9-10-22-27(5)15-13-23(31)26(3,4)21(27)12-16-29(22,30)7/h9,18-19,21-25,31-32H,10-17H2,1-8H3 |
|---|
| InChI Key | VJFLMYRRJUWADI-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as triterpenoids. These are terpene molecules containing six isoprene units. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Triterpenoids |
|---|
| Direct Parent | Triterpenoids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Triterpenoid
- Hydroxysteroid
- 12-hydroxysteroid
- Steroid
- Cyclic alcohol
- Secondary alcohol
- Organic oxygen compound
- Hydrocarbon derivative
- Organooxygen compound
- Alcohol
- Aliphatic homopolycyclic compound
|
|---|
| Molecular Framework | Aliphatic homopolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-004j-0026900000-525d926263177cd91663 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-00di-1011190000-68026ce195f4b0a2099f | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-004l-0000900000-98ea1b8bedbead647929 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-056r-1222900000-7f165506b3b4a15bfbfe | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0aor-8779200000-5877721a9b15f9fdff1f | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-004l-0000900000-98ea1b8bedbead647929 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-056r-1222900000-7f165506b3b4a15bfbfe | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0aor-8779200000-5877721a9b15f9fdff1f | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0006-0000900000-be67c01590579077e39c | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-006x-0000900000-19f0bfc1cf9b35c2ef2f | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a6r-0002900000-df10ca2f362dcd3ea4ea | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0006-0000900000-be67c01590579077e39c | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-006x-0000900000-19f0bfc1cf9b35c2ef2f | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a6r-0002900000-df10ca2f362dcd3ea4ea | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0006-0000900000-849799f7ece1fdb1b84b | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0006-0000900000-849799f7ece1fdb1b84b | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-000l-0000900000-f3fd398e566ceee628e2 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0006-0001900000-7bd975faf4f464c936d8 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0ffx-0983500000-a07179d837bfdca282e4 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00bi-1960000000-9ed3ea5b5ab829d22395 | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0035769 |
|---|
| FooDB ID | FDB014511 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | C00023426 |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | BREIN |
|---|
| Chemspider ID | 11343849 |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 12302049 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|