| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 01:15:26 UTC |
|---|
| Update Date | 2016-11-09 01:18:54 UTC |
|---|
| Accession Number | CHEM029577 |
|---|
| Identification |
|---|
| Common Name | Deacetylnomilin |
|---|
| Class | Small Molecule |
|---|
| Description | Isolated from orange (Citrus sinensis) and finger lime (Microcitrus australasica). Deacetylnomilin is found in lemon, sweet orange, and citrus. |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
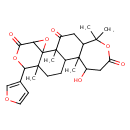
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Isolimonin | HMDB |
|
|---|
| Chemical Formula | C26H32O8 |
|---|
| Average Molecular Mass | 472.528 g/mol |
|---|
| Monoisotopic Mass | 472.210 g/mol |
|---|
| CAS Registry Number | 3264-90-2 |
|---|
| IUPAC Name | 7-(furan-3-yl)-13-hydroxy-1,8,12,17,17-pentamethyl-3,6,16-trioxapentacyclo[9.9.0.0²,⁴.0²,⁸.0¹²,¹⁸]icosane-5,15,20-trione |
|---|
| Traditional Name | 7-(furan-3-yl)-13-hydroxy-1,8,12,17,17-pentamethyl-3,6,16-trioxapentacyclo[9.9.0.0²,⁴.0²,⁸.0¹²,¹⁸]icosane-5,15,20-trione |
|---|
| SMILES | CC12CCC3C4(C)C(CC(=O)C3(C)C11OC1C(=O)OC2C1=COC=C1)C(C)(C)OC(=O)CC4O |
|---|
| InChI Identifier | InChI=1S/C26H32O8/c1-22(2)15-10-17(28)25(5)14(24(15,4)16(27)11-18(29)33-22)6-8-23(3)19(13-7-9-31-12-13)32-21(30)20-26(23,25)34-20/h7,9,12,14-16,19-20,27H,6,8,10-11H2,1-5H3 |
|---|
| InChI Key | HWAJASVMTDEFJN-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as limonoids. These are highly oxygenated, modified terpenoids with a prototypical structure either containing or derived from a precursor with a 4,4,8-trimethyl-17-furanylsteroid skeleton. All naturally occurring citrus limonoids contain a furan ring attached to the D-ring, at C-17, as well as oxygen containing functional groups at C-3, C-4, C-7, C-16 and C-17. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Triterpenoids |
|---|
| Direct Parent | Limonoids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Limonoid skeleton
- Naphthopyran
- Naphthalene
- Caprolactone
- 1,4-dioxepane
- Delta valerolactone
- Dioxepane
- Delta_valerolactone
- Oxepane
- Oxane
- Pyran
- Dicarboxylic acid or derivatives
- Heteroaromatic compound
- Furan
- Secondary alcohol
- Carboxylic acid ester
- Lactone
- Ketone
- Oxacycle
- Organoheterocyclic compound
- Carboxylic acid derivative
- Dialkyl ether
- Oxirane
- Ether
- Organic oxide
- Alcohol
- Hydrocarbon derivative
- Organic oxygen compound
- Carbonyl group
- Organooxygen compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-01si-2259800000-a4d140cdff7bea3a3998 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-003r-7064490000-0939502d59b9f57a162d | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0ab9-0001900000-ac2fd55de38221c65f6b | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a4u-0025900000-3428d6398664c9707925 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00kb-9416000000-5b63ee7c0bd320d2261e | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00b9-0001900000-7c933e17bd71d8b72217 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0umj-1006900000-28026242e599d86106f5 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4m-9107200000-389a14bbe7886e74cb64 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00di-0000900000-6e029cb28edf4d9b8aa3 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00di-0141900000-968227572d16a11da18f | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000i-2921200000-cd914e3f6b6a42b6f420 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00di-0000900000-6092626f870c8a12f250 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00di-0000900000-c9163d595834a7b09573 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-4013900000-8418108a5da482d487cc | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0035684 |
|---|
| FooDB ID | FDB014401 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | Not Available |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 13857953 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|