| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 01:15:23 UTC |
|---|
| Update Date | 2016-11-09 01:18:54 UTC |
|---|
| Accession Number | CHEM029576 |
|---|
| Identification |
|---|
| Common Name | Dukunolide A |
|---|
| Class | Small Molecule |
|---|
| Description | Constituent of the pollen of Helianthus annuus (sunflower). 3-Hydroxy-1-phenyl-1-hexadecanone is found in fats and oils. |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
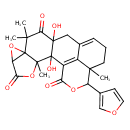
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 1-Benzoyl-2-pentadecanol | HMDB | | 1,1,1-trifluoro-2-Octanol | HMDB | | 1,1,1-Trifluorooctan-2-ol | HMDB |
|
|---|
| Chemical Formula | C26H26O9 |
|---|
| Average Molecular Mass | 482.479 g/mol |
|---|
| Monoisotopic Mass | 482.158 g/mol |
|---|
| CAS Registry Number | 97804-04-1 |
|---|
| IUPAC Name | 18-(furan-3-yl)-2,11-dihydroxy-3,9,9,17-tetramethyl-4,7,19-trioxahexacyclo[11.7.1.0²,¹¹.0³,⁸.0⁶,⁸.0¹⁷,²¹]henicosa-1(21),13-diene-5,10,20-trione |
|---|
| Traditional Name | 18-(furan-3-yl)-2,11-dihydroxy-3,9,9,17-tetramethyl-4,7,19-trioxahexacyclo[11.7.1.0²,¹¹.0³,⁸.0⁶,⁸.0¹⁷,²¹]henicosa-1(21),13-diene-5,10,20-trione |
|---|
| SMILES | CC12OC(=O)C3OC13C(C)(C)C(=O)C1(O)CC3=CCCC4(C)C(OC(=O)C(=C34)C21O)C1=COC=C1 |
|---|
| InChI Identifier | InChI=1S/C26H26O9/c1-21(2)20(29)24(30)10-12-6-5-8-22(3)14(12)15(18(27)33-16(22)13-7-9-32-11-13)25(24,31)23(4)26(21)17(34-26)19(28)35-23/h6-7,9,11,16-17,30-31H,5,8,10H2,1-4H3 |
|---|
| InChI Key | BATTZJIKLZSIAU-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as long-chain fatty alcohols. These are fatty alcohols that have an aliphatic tail of 13 to 21 carbon atoms. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Fatty Acyls |
|---|
| Sub Class | Fatty alcohols |
|---|
| Direct Parent | Long-chain fatty alcohols |
|---|
| Alternative Parents | |
|---|
| Substituents | - Alkyl-phenylketone
- Long chain fatty alcohol
- Butyrophenone
- Phenylketone
- Benzoyl
- Aryl alkyl ketone
- Aryl ketone
- Monocyclic benzene moiety
- Beta-hydroxy ketone
- Benzenoid
- Secondary alcohol
- Ketone
- Alcohol
- Hydrocarbon derivative
- Organic oxide
- Organic oxygen compound
- Organooxygen compound
- Aromatic homomonocyclic compound
|
|---|
| Molecular Framework | Aromatic homomonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0f6w-9187700000-0c2a7c908194de1bc5f3 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-002f-9500682000-d4a39e6552e42584c571 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_2) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_1) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS ("Dukunolide A,1TMS,#2" TMS) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-001i-0000900000-da8ac3e0960d9da6301a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00lr-1001900000-a7a4ec4957242dee0a6b | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0002-7709200000-399347021ab16db3424a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-001r-0000900000-bd77068acce7dc5d8ed7 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-01qi-1100900000-713c9a5813e43e4afe8b | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0aos-9806800000-317daa3f79d0b756a9b6 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-001i-0000900000-e224776f4657f4af7bc6 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-001i-2001900000-7d43705e485ecb09784a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-000i-2034900000-d50d05351d98039d86e2 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-001i-0000900000-0b0163d042220f372611 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-001i-1009800000-7a79187b060f7832fe21 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000y-2590500000-7d4e6f2c065482024976 | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0035677 |
|---|
| FooDB ID | FDB014393 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | Not Available |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 131751842 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|