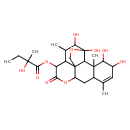| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-01di-6001900000-e1ce9105bdde68503ee1 | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-00ba-4520129000-40ed048370ebf98a64c3 | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_5) - 70eV, Positive | Not Available | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_4) - 70eV, Positive | Not Available | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_5) - 70eV, Positive | Not Available | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_6) - 70eV, Positive | Not Available | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_7) - 70eV, Positive | Not Available | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_8) - 70eV, Positive | Not Available | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_9) - 70eV, Positive | Not Available | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_10) - 70eV, Positive | Not Available | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_1) - 70eV, Positive | Not Available | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_2) - 70eV, Positive | Not Available | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_3) - 70eV, Positive | Not Available | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_4) - 70eV, Positive | Not Available | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_5) - 70eV, Positive | Not Available | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_6) - 70eV, Positive | Not Available | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_7) - 70eV, Positive | Not Available | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-004j-1104900000-7bb6188c59c74e7d63c3 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0umi-5109400000-decc9a710ceb50a6c12c | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00fr-6209000000-9a4e5b75b9a759c4c42d | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0f6t-1002900000-a716f8c5bf8de2ae5bad | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0101-9308800000-cada2918a5e89f56ba06 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00di-8109000000-017a1d307b0873e107bf | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0002-0000900000-f3195f31d3cb6feb3ab3 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-01t9-2009300000-8fcfd135168b4930c15d | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a6r-1109000000-00c65803bcdfcef153b4 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-1100900000-d51f5d15c51b1d4af910 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00mk-5509100000-ac5ed73af3ca8593505b | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004j-4009000000-bb012fc0dcf5c95a0d34 | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |