| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 01:12:02 UTC |
|---|
| Update Date | 2016-11-09 01:18:53 UTC |
|---|
| Accession Number | CHEM029495 |
|---|
| Identification |
|---|
| Common Name | Armexifolin |
|---|
| Class | Small Molecule |
|---|
| Description | Armexifolin is found in herbs and spices. Armexifolin is isolated from Tanacetum vulgare (tansy |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
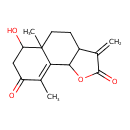
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 1-Epi-ludovicin C | HMDB | | 1-Epiludovicin C | HMDB | | [3AS-(3aalpha,5abeta,6beta,9bbeta)]-3a,5,5a,6,7,9b-hexahydro-6-hydroxy-5a,9-dimethyl-3-methylenenaphtho[1,2-b]furan-2,8(3H,4H)-dione | HMDB |
|
|---|
| Chemical Formula | C15H18O4 |
|---|
| Average Molecular Mass | 262.301 g/mol |
|---|
| Monoisotopic Mass | 262.121 g/mol |
|---|
| CAS Registry Number | 64929-15-3 |
|---|
| IUPAC Name | 6-hydroxy-5a,9-dimethyl-3-methylidene-2H,3H,3aH,4H,5H,5aH,6H,7H,8H,9bH-naphtho[1,2-b]furan-2,8-dione |
|---|
| Traditional Name | 6-hydroxy-5a,9-dimethyl-3-methylidene-3aH,4H,5H,6H,7H,9bH-naphtho[1,2-b]furan-2,8-dione |
|---|
| SMILES | CC1=C2C3OC(=O)C(=C)C3CCC2(C)C(O)CC1=O |
|---|
| InChI Identifier | InChI=1S/C15H18O4/c1-7-9-4-5-15(3)11(17)6-10(16)8(2)12(15)13(9)19-14(7)18/h9,11,13,17H,1,4-6H2,2-3H3 |
|---|
| InChI Key | QPXLDBMZJNDASA-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as eudesmanolides, secoeudesmanolides, and derivatives. These are terpenoids with a structure based on the eudesmanolide (a 3,5a,9-trimethyl-naphtho[1,2-b]furan-2-one derivative) or secoeudesmanolide (a 3,6-dimethyl-5-(pentan-2-yl)-1-benzofuran-2-one derivative) skeleton. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Terpene lactones |
|---|
| Direct Parent | Eudesmanolides, secoeudesmanolides, and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Eudesmanolide
- Sesquiterpenoid
- Naphthofuran
- Cyclohexenone
- Gamma butyrolactone
- Tetrahydrofuran
- Alpha,beta-unsaturated carboxylic ester
- Enoate ester
- Carboxylic acid ester
- Ketone
- Lactone
- Secondary alcohol
- Cyclic ketone
- Monocarboxylic acid or derivatives
- Carboxylic acid derivative
- Oxacycle
- Organoheterocyclic compound
- Hydrocarbon derivative
- Carbonyl group
- Organic oxide
- Organic oxygen compound
- Organooxygen compound
- Alcohol
- Aliphatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aliphatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-00xu-2980000000-4ca9118be40190ac58be | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-00xu-3192000000-c510f76cd3bda309dacf | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-01ot-0290000000-9b19184ee1cf2cf59c5e | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-01ot-0980000000-0c94b4b0092cd60afe9d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0gb9-7910000000-7f0551b78d28bce27f14 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03di-0090000000-d26fbae955b08a90028a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-03xu-0190000000-0ee5a1aa5d170c628a3b | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0uy1-2940000000-e07bcb34e59ebe720f5b | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03dl-0890000000-527d48fb87d02bc1e22d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0006-0940000000-9bb1c1b3c4db36220995 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000l-0910000000-baf1aa88cb99bdbeb1ee | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-02t9-0090000000-4fd8b99d59d84f8875b8 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-014i-0390000000-d5363cedd5bf3e29a571 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-3490000000-ce7839b7f577271afaff | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0035596 |
|---|
| FooDB ID | FDB014288 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | C00013062 |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 26503148 |
|---|
| ChEBI ID | 174461 |
|---|
| PubChem Compound ID | 13918461 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|