| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 01:11:51 UTC |
|---|
| Update Date | 2016-11-09 01:18:53 UTC |
|---|
| Accession Number | CHEM029491 |
|---|
| Identification |
|---|
| Common Name | 15-Acetoxyscirpene-3,4-diol 4-O-a-D-glucopyranoside |
|---|
| Class | Small Molecule |
|---|
| Description | From Fusarium sulphureum |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
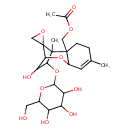
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (10'-Hydroxy-1',5'-dimethyl-11'-{[3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy}-8'-oxaspiro[oxirane-2,12'-tricyclo[7.2.1.0²,⁷]dodecan]-5'-en-2'-yl)methyl acetic acid | Generator |
|
|---|
| Chemical Formula | C23H34O11 |
|---|
| Average Molecular Mass | 486.510 g/mol |
|---|
| Monoisotopic Mass | 486.210 g/mol |
|---|
| CAS Registry Number | 99124-45-5 |
|---|
| IUPAC Name | 10'-hydroxy-1',5'-dimethyl-11'-{[3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy}-8'-oxaspiro[oxirane-2,12'-tricyclo[7.2.1.0²,⁷]dodecan]-5'-en-2'-ylmethyl acetate |
|---|
| Traditional Name | 10'-hydroxy-1',5'-dimethyl-11'-{[3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy}-8'-oxaspiro[oxirane-2,12'-tricyclo[7.2.1.0²,⁷]dodecan]-5'-en-2'-ylmethyl acetate |
|---|
| SMILES | CC(=O)OCC12CCC(C)=CC1OC1C(O)C(OC3OC(CO)C(O)C(O)C3O)C2(C)C11CO1 |
|---|
| InChI Identifier | InChI=1S/C23H34O11/c1-10-4-5-22(8-30-11(2)25)13(6-10)33-19-17(29)18(21(22,3)23(19)9-31-23)34-20-16(28)15(27)14(26)12(7-24)32-20/h6,12-20,24,26-29H,4-5,7-9H2,1-3H3 |
|---|
| InChI Key | HUIYHYRPRGTYKA-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as trichothecenes. These are sesquiterpene mycotoxins structurally characterized by the presence of an epoxide ring and a benzopyran derivative with a variant number of hydroxyl, acetyl, or other substituents. The most important structural features causing the biological activities of trichothecenes are the 12,13-epoxy ring, the presence of hydroxyl or acetyl groups at appropriate positions on the trichothecene nucleus and the structure and position of the side-chain. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Sesquiterpenoids |
|---|
| Direct Parent | Trichothecenes |
|---|
| Alternative Parents | Not Available |
|---|
| Substituents | Not Available |
|---|
| Molecular Framework | Aliphatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0aou-7563900000-1c0b4d4171730a89a2f7 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-02tc-8972108000-d064a0ffd2f496101bde | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00or-1027900000-e4a0b983baf1b84d415f | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-05r0-1589200000-99cb3f157319bbf1ce81 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-014i-5494000000-10dfd7a67aaf90279ab0 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-007c-5206900000-3a7f31121457db3b0a00 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0abc-7429400000-d43dc4cfa2dd40a071b9 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-2900000000-23e378633777d830c744 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000f-1000900000-cbe4cf401eb5c8821200 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4i-9000700000-254367a93a64c280b1f2 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-9003200000-e239a135ebd1e635d5dc | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-004i-0000900000-ff6a6b1c4250cfdec848 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-004v-1020900000-41889cb27f2232559312 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-052e-9501100000-60654292a029eac5d868 | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0035590 |
|---|
| FooDB ID | FDB014282 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | Not Available |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 74105490 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|