| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 01:01:39 UTC |
|---|
| Update Date | 2016-11-09 01:18:50 UTC |
|---|
| Accession Number | CHEM029230 |
|---|
| Identification |
|---|
| Common Name | Cucurbitaxanthin B |
|---|
| Class | Small Molecule |
|---|
| Description | Cucurbitaxanthin B is found in fruits. Cucurbitaxanthin B is isolated from pumpkin (Cucurbita maxima) and from paprika fruits. |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
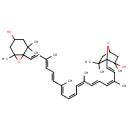
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (3S,3's,5R,5'r,6R,6's)-3,6:4',5'-Diepoxy-5,5',6,6'-tetrahydro-3',5-dihydroxy-beta,beta-carotene | HMDB | | 3,6:5',6'-Diepoxy-5,5',6,6'-tetrahydro-b,b-carotene-3',5-diol | HMDB |
|
|---|
| Chemical Formula | C40H56O4 |
|---|
| Average Molecular Mass | 600.870 g/mol |
|---|
| Monoisotopic Mass | 600.418 g/mol |
|---|
| CAS Registry Number | 107390-61-4 |
|---|
| IUPAC Name | 1-[(1E,3Z,5E,7Z,9Z,11E,13E,15E,17E)-18-{4-hydroxy-2,2,6-trimethyl-7-oxabicyclo[4.1.0]heptan-1-yl}-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl]-2,6,6-trimethyl-7-oxabicyclo[2.2.1]heptan-2-ol |
|---|
| Traditional Name | 1-[(1E,3Z,5E,7Z,9Z,11E,13E,15E,17E)-18-{4-hydroxy-2,2,6-trimethyl-7-oxabicyclo[4.1.0]heptan-1-yl}-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl]-2,6,6-trimethyl-7-oxabicyclo[2.2.1]heptan-2-ol |
|---|
| SMILES | C\C(\C=C\C=C(/C)\C=C\C12OC1(C)CC(O)CC2(C)C)=C/C=C\C=C(\C)/C=C/C=C(/C)\C=C\C12OC(CC1(C)C)CC2(C)O |
|---|
| InChI Identifier | InChI=1S/C40H56O4/c1-29(17-13-19-31(3)21-23-39-36(7,8)27-34(43-39)28-37(39,9)42)15-11-12-16-30(2)18-14-20-32(4)22-24-40-35(5,6)25-33(41)26-38(40,10)44-40/h11-24,33-34,41-42H,25-28H2,1-10H3/b12-11-,17-13+,18-14+,23-21+,24-22+,29-15-,30-16+,31-19-,32-20+ |
|---|
| InChI Key | WBXYNQBROQPCES-CIGGGVHVSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as xanthophylls. These are carotenoids containing an oxygenated carotene backbone. Carotenes are characterized by the presence of two end-groups (mostly cyclohexene rings, but also cyclopentene rings or acyclic groups) linked by a long branched alkyl chain. Carotenes belonging form a subgroup of the carotenoids family. Xanthophylls arise by oxygenation of the carotene backbone. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Tetraterpenoids |
|---|
| Direct Parent | Xanthophylls |
|---|
| Alternative Parents | |
|---|
| Substituents | - Xanthophyll
- Oxepane
- Monosaccharide
- Tetrahydrofuran
- Tertiary alcohol
- Cyclic alcohol
- Secondary alcohol
- Oxacycle
- Organoheterocyclic compound
- Ether
- Oxirane
- Dialkyl ether
- Organic oxygen compound
- Alcohol
- Organooxygen compound
- Hydrocarbon derivative
- Aliphatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aliphatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-052r-2000090000-343a479313d129be9f4b | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-0a4i-4200009000-0f84f31ee8495762f0e5 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_1) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_2) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_1) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS ("Cucurbitaxanthin B,1TMS,#1" TMS) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-001i-0175895000-a25d198810836f20c34c | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03e9-0395310000-537b327ef86b0752210a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-08fs-3492100000-ee29fc153dbb80224d69 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-0100090000-cae1ae6eb80b22f1aa4b | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-000t-0100090000-6146b314bf922fbcacf1 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0pbj-2900360000-7193bc86a2637a3152dc | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0ue9-0447297000-c7593087db87e1c06d65 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0gc0-1223292000-8ba93b79ae9933368f64 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-003j-1924210000-701c28c18f42ceab27b5 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-0400090000-a00cc9954b33b66e1315 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0002-0203290000-945000aad4eb7905d920 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-000b-0537190000-70c019e27773431ad662 | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0035320 |
|---|
| FooDB ID | FDB013990 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | C00022954 |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 35013903 |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 131751705 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|