| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 01:01:35 UTC |
|---|
| Update Date | 2016-11-09 01:18:50 UTC |
|---|
| Accession Number | CHEM029229 |
|---|
| Identification |
|---|
| Common Name | Cucurbitaxanthin A |
|---|
| Class | Small Molecule |
|---|
| Description | Cucurbitaxanthin A is found in fruits. Cucurbitaxanthin A is a constituent of pumpkin (Cucurbita maxima) and of paprika (Capsicum annuum var. longum) fruits. |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
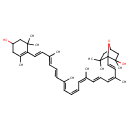
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (3S,3'r,5R,6R)-3,6-Epoxy-5,6-dihydro-3',5-dihydroxy-beta,beta-carotene | HMDB | | 3,6-Epoxy-5,6-dihydro-b,b-carotene-3',5-diol | HMDB | | Zeaxanthin 3,6-epoxide (incorr.) | HMDB | | Cucurbitaxanthin a | MeSH |
|
|---|
| Chemical Formula | C40H56O3 |
|---|
| Average Molecular Mass | 584.871 g/mol |
|---|
| Monoisotopic Mass | 584.423 g/mol |
|---|
| CAS Registry Number | 103955-77-7 |
|---|
| IUPAC Name | 1-[(1E,3Z,5E,7Z,9Z,11E,13E,15E,17E)-18-(4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl)-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl]-2,6,6-trimethyl-7-oxabicyclo[2.2.1]heptan-2-ol |
|---|
| Traditional Name | 1-[(1E,3Z,5E,7Z,9Z,11E,13E,15E,17E)-18-(4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl)-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl]-2,6,6-trimethyl-7-oxabicyclo[2.2.1]heptan-2-ol |
|---|
| SMILES | C\C(\C=C\C=C(/C)\C=C\C1=C(C)CC(O)CC1(C)C)=C/C=C\C=C(\C)/C=C/C=C(/C)\C=C\C12OC(CC1(C)C)CC2(C)O |
|---|
| InChI Identifier | InChI=1S/C40H56O3/c1-29(17-13-19-31(3)21-22-36-33(5)25-34(41)26-37(36,6)7)15-11-12-16-30(2)18-14-20-32(4)23-24-40-38(8,9)27-35(43-40)28-39(40,10)42/h11-24,34-35,41-42H,25-28H2,1-10H3/b12-11-,17-13+,18-14+,22-21+,24-23+,29-15+,30-16-,31-19+,32-20- |
|---|
| InChI Key | LMIFPRVTIOZTJN-NYORYHGYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as xanthophylls. These are carotenoids containing an oxygenated carotene backbone. Carotenes are characterized by the presence of two end-groups (mostly cyclohexene rings, but also cyclopentene rings or acyclic groups) linked by a long branched alkyl chain. Carotenes belonging form a subgroup of the carotenoids family. Xanthophylls arise by oxygenation of the carotene backbone. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Tetraterpenoids |
|---|
| Direct Parent | Xanthophylls |
|---|
| Alternative Parents | |
|---|
| Substituents | - Xanthophyll
- Monosaccharide
- Tetrahydrofuran
- Tertiary alcohol
- Cyclic alcohol
- Secondary alcohol
- Oxacycle
- Organoheterocyclic compound
- Ether
- Dialkyl ether
- Organic oxygen compound
- Hydrocarbon derivative
- Organooxygen compound
- Alcohol
- Aliphatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aliphatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-014i-1000090000-b5bad558c87182a08aaf | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-052f-6000039000-14a671f132447857f072 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS ("Cucurbitaxanthin A,1TMS,#1" TMS) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_1) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_2) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_1) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014i-0112490000-b536673bf9530e236361 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03yi-0776940000-c6b1fb3127704a181aaf | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03di-4794110000-c6ff1c636412bbaffade | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-001i-0000090000-dd2709b5e48ac1604ea3 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00lr-0100090000-ead60f990571de6a5f16 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0pvr-3900260000-101603894e6c9939344f | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-001i-0000090000-b3c3d9eef7b0427b1314 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-001i-0113190000-0e2857a40c9434199bf0 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-014l-0229420000-dc28d71a47f878b945be | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-0355690000-05ca4962beb7a4a003be | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-014i-0727590000-a031b89bd48b36d04c56 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0ap1-1889100000-89484715e153f8ab5a41 | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0035319 |
|---|
| FooDB ID | FDB013989 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | C00022948 |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 35013902 |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 131751704 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|