| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 00:57:49 UTC |
|---|
| Update Date | 2016-11-09 01:18:49 UTC |
|---|
| Accession Number | CHEM029146 |
|---|
| Identification |
|---|
| Common Name | (3beta,17alpha,23S)-17,23-Epoxy-3,28,29-trihydroxy-27-norlanost-8-en-24-one |
|---|
| Class | Small Molecule |
|---|
| Description | (3beta,17alpha,23S)-17,23-Epoxy-3,28,29-trihydroxy-27-norlanost-8-en-24-one is found in herbs and spices. Aglycone from Muscari comosum (tassel hyacinth). |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
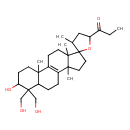
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (3b,17a,23S)-17,23-Epoxy-3,28,29-trihydroxy-27-norlanost-8-en-24-one | Generator | | (3Β,17α,23S)-17,23-epoxy-3,28,29-trihydroxy-27-norlanost-8-en-24-one | Generator |
|
|---|
| Chemical Formula | C29H46O5 |
|---|
| Average Molecular Mass | 474.673 g/mol |
|---|
| Monoisotopic Mass | 474.335 g/mol |
|---|
| CAS Registry Number | Not Available |
|---|
| IUPAC Name | 1-[5'-hydroxy-6',6'-bis(hydroxymethyl)-2',3,11',15'-tetramethylspiro[oxolane-2,14'-tetracyclo[8.7.0.0²,⁷.0¹¹,¹⁵]heptadecan]-1'(10')-en-5-yl]propan-1-one |
|---|
| Traditional Name | 1-[5'-hydroxy-6',6'-bis(hydroxymethyl)-2',3,11',15'-tetramethylspiro[oxolane-2,14'-tetracyclo[8.7.0.0²,⁷.0¹¹,¹⁵]heptadecan]-1'(10')-en-5-yl]propan-1-one |
|---|
| SMILES | CCC(=O)C1CC(C)C2(CCC3(C)C4=C(CCC23C)C2(C)CCC(O)C(CO)(CO)C2CC4)O1 |
|---|
| InChI Identifier | InChI=1S/C29H46O5/c1-6-21(32)22-15-18(2)29(34-22)14-13-26(4)20-7-8-23-25(3,19(20)9-12-27(26,29)5)11-10-24(33)28(23,16-30)17-31/h18,22-24,30-31,33H,6-17H2,1-5H3 |
|---|
| InChI Key | PGCCXLDWXXFVMP-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as triterpenoids. These are terpene molecules containing six isoprene units. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Triterpenoids |
|---|
| Direct Parent | Triterpenoids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Triterpenoid
- 3-hydroxysteroid
- Hydroxysteroid
- Steroid
- Cyclic alcohol
- Tetrahydrofuran
- Ketone
- Secondary alcohol
- Dialkyl ether
- Ether
- Oxacycle
- Organoheterocyclic compound
- Hydrocarbon derivative
- Alcohol
- Organic oxygen compound
- Organic oxide
- Organooxygen compound
- Primary alcohol
- Carbonyl group
- Aliphatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aliphatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0a4j-2003900000-83c403f8d3c2f203a293 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (3 TMS) - 70eV, Positive | splash10-01t9-3100019000-7de4099e52ec6703a3c2 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0a4i-0002900000-323021f5e2f09c93f492 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a4r-1023900000-bc006ebeef1a457682bc | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000i-0049000000-f6d5fa832338fa134192 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00di-0000900000-0fa0f1bfbf107cc2aa1c | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0adl-0000900000-d75ca516b55bb1191e8e | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-052r-2009300000-fc13b34f9b9709f8dbc8 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-004i-0001900000-aba354fdc291a801c6c9 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00p1-0234900000-005f3badb1b04de269a3 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0006-9256400000-ebb1b9a73137af69a75a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00di-0000900000-221edc4136d4fc255e65 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-000i-1009200000-7be6ca4dbaade3e6f01a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-03di-0000900000-2366b5e46ef4d0e98cba | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0035221 |
|---|
| FooDB ID | FDB013870 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | C00010787 |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | Not Available |
|---|
| ChEBI ID | 167967 |
|---|
| PubChem Compound ID | 73816066 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|