| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 00:57:06 UTC |
|---|
| Update Date | 2016-11-09 01:18:49 UTC |
|---|
| Accession Number | CHEM029129 |
|---|
| Identification |
|---|
| Common Name | Yucalexin B5 |
|---|
| Class | Small Molecule |
|---|
| Description | Yucalexin B5 is found in root vegetables. Yucalexin B5 is a constituent of cassava roots Manihot esculenta. |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
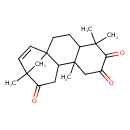
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| ent-15-Beyerene-2,3,12-trione | HMDB | | ent-2-Hydroxy-1,15-beyeradiene-3,12-dione | HMDB |
|
|---|
| Chemical Formula | C20H26O3 |
|---|
| Average Molecular Mass | 314.419 g/mol |
|---|
| Monoisotopic Mass | 314.188 g/mol |
|---|
| CAS Registry Number | 50719-31-8 |
|---|
| IUPAC Name | 5,5,9,13-tetramethyltetracyclo[11.2.1.0¹,¹⁰.0⁴,⁹]hexadec-14-ene-6,7,12-trione |
|---|
| Traditional Name | 5,5,9,13-tetramethyltetracyclo[11.2.1.0¹,¹⁰.0⁴,⁹]hexadec-14-ene-6,7,12-trione |
|---|
| SMILES | CC12CC3(CCC4C(C)(C)C(=O)C(=O)CC4(C)C3CC1=O)C=C2 |
|---|
| InChI Identifier | InChI=1S/C20H26O3/c1-17(2)13-5-6-20-8-7-18(3,11-20)15(22)9-14(20)19(13,4)10-12(21)16(17)23/h7-8,13-14H,5-6,9-11H2,1-4H3 |
|---|
| InChI Key | KYOFWFWEZCLKDW-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as diterpenoids. These are terpene compounds formed by four isoprene units. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Diterpenoids |
|---|
| Direct Parent | Diterpenoids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Diterpenoid
- Beyerane diterpenoid
- Cyclic ketone
- Ketone
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Carbonyl group
- Aliphatic homopolycyclic compound
|
|---|
| Molecular Framework | Aliphatic homopolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-00dr-0190000000-355867c9a2c1d9d09de4 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014i-0169000000-2287f3e574bb058a4bad | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-014j-0493000000-82281229f281e0c82ec6 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-053r-3960000000-487aa64a413a4412a429 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03di-0009000000-0437fc7330157ef9afc9 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-03di-0029000000-98113a9e7ffb7fe7310f | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-000w-4090000000-c30ac72150b8711fa5c0 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03di-0009000000-d8638daf32922c227464 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-03di-0059000000-1e0ad665179afebed57f | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-03dj-0094000000-8fba71c40c17f0f31759 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014i-0039000000-91283a7dc0db4604ceb7 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0frb-0292000000-a71a0e48cf25ca4df87e | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0f72-3890000000-f66c59810a3beeeb2370 | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0035200 |
|---|
| FooDB ID | FDB013846 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 35013872 |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 131751681 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|