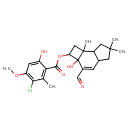| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 00:54:20 UTC |
|---|
| Update Date | 2016-11-09 01:18:49 UTC |
|---|
| Accession Number | CHEM029072 |
|---|
| Identification |
|---|
| Common Name | Armillaridin |
|---|
| Class | Small Molecule |
|---|
| Description | Armillaridin is found in mushrooms. Armillaridin is from Armillaria mellea (honey mushroom |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 3-Formyl-2a-hydroxy-6,6,7b-trimethyl-1H,2H,2ah,4ah,5H,6H,7H,7ah,7BH-cyclobuta[e]inden-2-yl 3-chloro-6-hydroxy-4-methoxy-2-methylbenzoic acid | HMDB | | Armillaridin | MeSH |
|
|---|
| Chemical Formula | C24H29ClO6 |
|---|
| Average Molecular Mass | 448.936 g/mol |
|---|
| Monoisotopic Mass | 448.165 g/mol |
|---|
| CAS Registry Number | 96684-80-9 |
|---|
| IUPAC Name | 3-formyl-2a-hydroxy-6,6,7b-trimethyl-1H,2H,2aH,4aH,5H,6H,7H,7aH,7bH-cyclobuta[e]inden-2-yl 3-chloro-6-hydroxy-4-methoxy-2-methylbenzoate |
|---|
| Traditional Name | 3-formyl-2a-hydroxy-6,6,7b-trimethyl-1H,2H,4aH,5H,7H,7aH-cyclobuta[e]inden-2-yl 3-chloro-6-hydroxy-4-methoxy-2-methylbenzoate |
|---|
| SMILES | COC1=C(Cl)C(C)=C(C(=O)OC2CC3(C)C4CC(C)(C)CC4C=C(C=O)C23O)C(O)=C1 |
|---|
| InChI Identifier | InChI=1S/C24H29ClO6/c1-12-19(16(27)7-17(30-5)20(12)25)21(28)31-18-10-23(4)15-9-22(2,3)8-13(15)6-14(11-26)24(18,23)29/h6-7,11,13,15,18,27,29H,8-10H2,1-5H3 |
|---|
| InChI Key | QETHRCCHQRWBIJ-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as melleolides and analogues. Melleolides and analogues are compounds with a structure characterized by the presence of a 2-hydroxy-4-methoxy-6-methylbenzoic acid derivative linked to a 3,6,6,7b-tetramethyl-cyclobuta[e]indene moiety. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Sesquiterpenoids |
|---|
| Direct Parent | Melleolides and analogues |
|---|
| Alternative Parents | |
|---|
| Substituents | - Melleolide-skeleton
- O-hydroxybenzoic acid ester
- P-methoxybenzoic acid or derivatives
- Methoxyphenol
- Benzoate ester
- Salicylic acid or derivatives
- 3-halobenzoic acid or derivatives
- Halobenzoic acid or derivatives
- Benzoic acid or derivatives
- Benzoyl
- 4-halophenol
- Phenoxy compound
- M-cresol
- Anisole
- 4-chlorophenol
- Phenol ether
- Methoxybenzene
- Chlorobenzene
- Halobenzene
- Alkyl aryl ether
- 1-hydroxy-2-unsubstituted benzenoid
- Phenol
- Toluene
- Aryl halide
- Monocyclic benzene moiety
- Aryl chloride
- Benzenoid
- Tertiary alcohol
- Vinylogous acid
- Cyclobutanol
- Carboxylic acid ester
- Monocarboxylic acid or derivatives
- Ether
- Carboxylic acid derivative
- Alcohol
- Organohalogen compound
- Organochloride
- Organooxygen compound
- Hydrocarbon derivative
- Organic oxide
- Aldehyde
- Carbonyl group
- Organic oxygen compound
- Aromatic homopolycyclic compound
|
|---|
| Molecular Framework | Aromatic homopolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0a4j-2690000000-d1e2a782cae06dcb0016 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-004i-2090010000-20879ec1fdf21785cd7d | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0002-0340900000-3f3ccad340e6a74edd38 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000t-1980400000-3a88f59cdddfd3cb666b | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00kb-2920000000-bdab1fb165eaf2b8a220 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-0230900000-72fd4909e00e753b4a0a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-006t-0890800000-a9fab9919d5faa334943 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00di-1940000000-f7446247539fd3204dbe | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-0110900000-5414efd65222af117874 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00xs-1890600000-85b50da1aad079e7c91a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-059t-8920300000-1fbcfdfc74264fa692f0 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0002-0270900000-6bd11b01d197d22444ea | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0002-1942100000-b8139a5a5531057d9ce3 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0002-8983000000-e6a86b002cef7e8982ac | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0035134 |
|---|
| FooDB ID | FDB013770 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | C00021454 |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 35013853 |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 75629634 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|