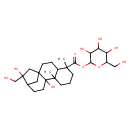| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 00:52:49 UTC |
|---|
| Update Date | 2016-11-09 01:18:48 UTC |
|---|
| Accession Number | CHEM029038 |
|---|
| Identification |
|---|
| Common Name | Cofaryloside |
|---|
| Class | Small Molecule |
|---|
| Description | Constituent of green coffee beans. Cofaryloside is found in coffee and coffee products. |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Lithocholic acid, methyl ester | HMDB | | Methyl 3alpha-hydroxy-5beta-cholan-24-Oate | HMDB | | Methyl lithocholate | HMDB | | 3,4,5-Trihydroxy-6-(hydroxymethyl)oxan-2-yl 10,14-dihydroxy-14-(hydroxymethyl)-5,9-dimethyltetracyclo[11.2.1.0¹,¹⁰.0⁴,⁹]hexadecane-5-carboxylic acid | Generator |
|
|---|
| Chemical Formula | C26H42O10 |
|---|
| Average Molecular Mass | 514.606 g/mol |
|---|
| Monoisotopic Mass | 514.278 g/mol |
|---|
| CAS Registry Number | 63558-43-0 |
|---|
| IUPAC Name | 3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl 10,14-dihydroxy-14-(hydroxymethyl)-5,9-dimethyltetracyclo[11.2.1.0¹,¹⁰.0⁴,⁹]hexadecane-5-carboxylate |
|---|
| Traditional Name | 3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl 10,14-dihydroxy-14-(hydroxymethyl)-5,9-dimethyltetracyclo[11.2.1.0¹,¹⁰.0⁴,⁹]hexadecane-5-carboxylate |
|---|
| SMILES | CC1(CCCC2(C)C1CCC13CC(CCC21O)C(O)(CO)C3)C(=O)OC1OC(CO)C(O)C(O)C1O |
|---|
| InChI Identifier | InChI=1S/C26H42O10/c1-22(21(32)36-20-19(31)18(30)17(29)15(11-27)35-20)6-3-7-23(2)16(22)5-8-24-10-14(4-9-26(23,24)34)25(33,12-24)13-28/h14-20,27-31,33-34H,3-13H2,1-2H3 |
|---|
| InChI Key | VEZJUFUSLIUNOY-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as diterpene glycosides. These are diterpenoids in which an isoprene unit is glycosylated. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Terpene glycosides |
|---|
| Direct Parent | Diterpene glycosides |
|---|
| Alternative Parents | |
|---|
| Substituents | - Diterpene glycoside
- Diterpenoid
- Kaurane diterpenoid
- Hexose monosaccharide
- Monosaccharide
- Oxane
- Cyclic alcohol
- Tertiary alcohol
- Carboxylic acid ester
- Secondary alcohol
- Polyol
- Organoheterocyclic compound
- Oxacycle
- Monocarboxylic acid or derivatives
- Acetal
- Carboxylic acid derivative
- Primary alcohol
- Organooxygen compound
- Hydrocarbon derivative
- Organic oxide
- Organic oxygen compound
- Carbonyl group
- Alcohol
- Aliphatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aliphatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-008j-3401900000-91188b84868934763b45 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-05fu-9441116000-4074d3fd5f6c1c74bd05 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0frj-0209310000-739970a4f0166f9c8c2d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-0219200000-e506957aa4b5e85acc4c | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000i-3559100000-8241df05d19dda33f559 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0il0-0109330000-7a60523fdb6d7d39fe2a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0h9r-2309300000-32361b19904e17979330 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0znc-7109000000-7180ecadf02bea99e169 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03di-0002090000-764df8c28fa16d24a6ac | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-03di-3307390000-862cf6d6c3d52bdf5586 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0pb9-7109010000-ff98f5156b461afb6d9d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014i-0103390000-3f8640ab76a9da78aa42 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00rj-1609210000-a3dbaa110cd9b6449e27 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4i-9212010000-05e549aea342add32e97 | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0035096 |
|---|
| FooDB ID | FDB013725 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | Not Available |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 131751664 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|