| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 00:38:38 UTC |
|---|
| Update Date | 2016-11-09 01:18:44 UTC |
|---|
| Accession Number | CHEM028713 |
|---|
| Identification |
|---|
| Common Name | Glabric acid |
|---|
| Class | Small Molecule |
|---|
| Description | Glabric acid is found in herbs and spices. Glabric acid is isolated from Glycyrrhiza glabra (licorice |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
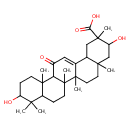
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Glabrate | Generator | | 3,10-Dihydroxy-2,4a,6a,6b,9,9,12a-heptamethyl-13-oxo-1,2,3,4,4a,5,6,6a,6b,7,8,8a,9,10,11,12,12a,12b,13,14b-icosahydropicene-2-carboxylate | HMDB |
|
|---|
| Chemical Formula | C30H46O5 |
|---|
| Average Molecular Mass | 486.683 g/mol |
|---|
| Monoisotopic Mass | 486.335 g/mol |
|---|
| CAS Registry Number | 22327-86-2 |
|---|
| IUPAC Name | 3,10-dihydroxy-2,4a,6a,6b,9,9,12a-heptamethyl-13-oxo-1,2,3,4,4a,5,6,6a,6b,7,8,8a,9,10,11,12,12a,12b,13,14b-icosahydropicene-2-carboxylic acid |
|---|
| Traditional Name | 3,10-dihydroxy-2,4a,6a,6b,9,9,12a-heptamethyl-13-oxo-3,4,5,6,7,8,8a,10,11,12,12b,14b-dodecahydro-1H-picene-2-carboxylic acid |
|---|
| SMILES | CC1(C)C(O)CCC2(C)C1CCC1(C)C2C(=O)C=C2C3CC(C)(C(O)CC3(C)CCC12C)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C30H46O5/c1-25(2)20-8-11-30(7)23(27(20,4)10-9-21(25)32)19(31)14-17-18-15-28(5,24(34)35)22(33)16-26(18,3)12-13-29(17,30)6/h14,18,20-23,32-33H,8-13,15-16H2,1-7H3,(H,34,35) |
|---|
| InChI Key | HFTWTHSIMCSLFQ-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as triterpenoids. These are terpene molecules containing six isoprene units. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Triterpenoids |
|---|
| Direct Parent | Triterpenoids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Triterpenoid
- Beta-hydroxy acid
- Cyclohexenone
- Hydroxy acid
- Cyclic alcohol
- Ketone
- Secondary alcohol
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Carboxylic acid derivative
- Alcohol
- Hydrocarbon derivative
- Organic oxide
- Carbonyl group
- Organic oxygen compound
- Organooxygen compound
- Aliphatic homopolycyclic compound
|
|---|
| Molecular Framework | Aliphatic homopolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-05tb-0209700000-4cb71c2e5d17e366436a | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-014i-1020917000-a88fc6253899627c5679 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0gb9-0000900000-4168721ccc7d1399e77b | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0uxr-0001900000-0519aa306678e311bbec | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0gi9-1419700000-7a86613b1e680692dc40 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000i-0000900000-9b43b74461515be63429 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00y3-0000900000-a442279bc1aa463e8431 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00b9-0000900000-e2da884754cda83a5289 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-0000900000-02676053c3c408d832e9 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00ri-0002900000-22a3aa4119c332747d93 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-014i-8849300000-c3668ad0960dfc3a53cd | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000i-0000900000-6a75a140e72d68a6c193 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-000i-0000900000-30217b326d2001e5e5de | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0ktr-1000900000-2d4db8fc50a63bddb9ff | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0034689 |
|---|
| FooDB ID | FDB013219 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | C00033956 |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 35013773 |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 75144805 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|