| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 00:32:02 UTC |
|---|
| Update Date | 2016-11-09 01:18:43 UTC |
|---|
| Accession Number | CHEM028551 |
|---|
| Identification |
|---|
| Common Name | Queretaroic acid |
|---|
| Class | Small Molecule |
|---|
| Description | Queretaroic acid is found in cereals and cereal products. Queretaroic acid is a constituent of Chenopodium quinoa (quinoa), Stenocereus thurberi (organ pipe cactus) |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
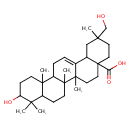
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Queretaroate | Generator | | 10-Hydroxy-2-(hydroxymethyl)-2,6a,6b,9,9,12a-hexamethyl-1,2,3,4,4a,5,6,6a,6b,7,8,8a,9,10,11,12,12a,12b,13,14b-icosahydropicene-4a-carboxylate | HMDB |
|
|---|
| Chemical Formula | C30H48O4 |
|---|
| Average Molecular Mass | 472.700 g/mol |
|---|
| Monoisotopic Mass | 472.355 g/mol |
|---|
| CAS Registry Number | 511-82-0 |
|---|
| IUPAC Name | 10-hydroxy-2-(hydroxymethyl)-2,6a,6b,9,9,12a-hexamethyl-1,2,3,4,4a,5,6,6a,6b,7,8,8a,9,10,11,12,12a,12b,13,14b-icosahydropicene-4a-carboxylic acid |
|---|
| Traditional Name | 10-hydroxy-2-(hydroxymethyl)-2,6a,6b,9,9,12a-hexamethyl-1,3,4,5,6,7,8,8a,10,11,12,12b,13,14b-tetradecahydropicene-4a-carboxylic acid |
|---|
| SMILES | CC1(C)C(O)CCC2(C)C1CCC1(C)C2CC=C2C3CC(C)(CO)CCC3(CCC12C)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C30H48O4/c1-25(2)21-9-12-29(6)22(27(21,4)11-10-23(25)32)8-7-19-20-17-26(3,18-31)13-15-30(20,24(33)34)16-14-28(19,29)5/h7,20-23,31-32H,8-18H2,1-6H3,(H,33,34) |
|---|
| InChI Key | CZWBKSDPBWNHGO-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as triterpenoids. These are terpene molecules containing six isoprene units. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Triterpenoids |
|---|
| Direct Parent | Triterpenoids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Triterpenoid
- Cyclic alcohol
- Secondary alcohol
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Carboxylic acid derivative
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Primary alcohol
- Organooxygen compound
- Carbonyl group
- Alcohol
- Aliphatic homopolycyclic compound
|
|---|
| Molecular Framework | Aliphatic homopolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-059f-0014900000-e4c4cc7889c2dd670450 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (3 TMS) - 70eV, Positive | splash10-00di-1000049000-ebf2031e1ad0333374be | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0a4i-0000900000-6ab12dfeea8d967e606d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a4r-0001900000-a8eb7591d0723ddc1dcf | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a59-4149800000-d5932048258cd7ab193a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00di-0000900000-c54866b503ba4e7ab68e | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0kor-0001900000-f42fcc48adbfd9271859 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0002-1005900000-4be075a08e6b5f3623c6 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00di-0000900000-a425355fd8f68e6dc919 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0avi-1202900000-7808b66fc425ed4c3e6c | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-053j-1955000000-e115a19bff6497045cde | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00di-0000900000-ce83f241675374237da4 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00di-0000900000-0c83e360efa63bcacca4 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0fk9-0001900000-3fc17774ca968db5d368 | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0034523 |
|---|
| FooDB ID | FDB013024 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | C00031116 |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 4477681 |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 5319375 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|