| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 00:20:41 UTC |
|---|
| Update Date | 2016-11-09 01:18:39 UTC |
|---|
| Accession Number | CHEM028277 |
|---|
| Identification |
|---|
| Common Name | (3beta,4alpha,5alpha,24(28)Z)-4,14-Dimethylstigmasta-9(11),24(28)-dien-3-ol |
|---|
| Class | Small Molecule |
|---|
| Description | (3beta,4alpha,5alpha,24(28)Z)-4,14-Dimethylstigmasta-9(11),24(28)-dien-3-ol is found in pulses. (3beta,4alpha,5alpha,24(28)Z)-4,14-Dimethylstigmasta-9(11),24(28)-dien-3-ol is a constituent of Phaseolus vulgaris (kidney bean). |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
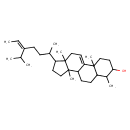
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (3b,4a,5a,24(28)Z)-4,14-Dimethylstigmasta-9(11),24(28)-dien-3-ol | Generator | | (3Β,4α,5α,24(28)Z)-4,14-dimethylstigmasta-9(11),24(28)-dien-3-ol | Generator |
|
|---|
| Chemical Formula | C31H52O |
|---|
| Average Molecular Mass | 440.744 g/mol |
|---|
| Monoisotopic Mass | 440.402 g/mol |
|---|
| CAS Registry Number | 123086-83-9 |
|---|
| IUPAC Name | 2,6,11,15-tetramethyl-14-[(5Z)-5-(propan-2-yl)hept-5-en-2-yl]tetracyclo[8.7.0.0²,⁷.0¹¹,¹⁵]heptadec-1(17)-en-5-ol |
|---|
| Traditional Name | 14-[(5Z)-5-isopropylhept-5-en-2-yl]-2,6,11,15-tetramethyltetracyclo[8.7.0.0²,⁷.0¹¹,¹⁵]heptadec-1(17)-en-5-ol |
|---|
| SMILES | C\C=C(\CCC(C)C1CCC2(C)C3CCC4C(C)C(O)CCC4(C)C3=CCC12C)C(C)C |
|---|
| InChI Identifier | InChI=1S/C31H52O/c1-9-23(20(2)3)11-10-21(4)24-14-18-31(8)27-13-12-25-22(5)28(32)16-17-29(25,6)26(27)15-19-30(24,31)7/h9,15,20-22,24-25,27-28,32H,10-14,16-19H2,1-8H3/b23-9- |
|---|
| InChI Key | ZIZODWXWMZVEEM-AQHIEDMUSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as stigmastanes and derivatives. These are sterol lipids with a structure based on the stigmastane skeleton, which consists of a cholestane moiety bearing an ethyl group at the carbon atom C24. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Steroids and steroid derivatives |
|---|
| Sub Class | Stigmastanes and derivatives |
|---|
| Direct Parent | Stigmastanes and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Triterpenoid
- Stigmastane-skeleton
- Hydroxysteroid
- 3-hydroxysteroid
- Cyclic alcohol
- Secondary alcohol
- Organic oxygen compound
- Hydrocarbon derivative
- Organooxygen compound
- Alcohol
- Aliphatic homopolycyclic compound
|
|---|
| Molecular Framework | Aliphatic homopolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-03fr-1005900000-afb7d9f37bb78c7eea31 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-000t-2001900000-bf4d037ea9deb1cf3785 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00dl-0002900000-f102a52b6d16b0a8e12a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0095-5229300000-1a2fc432f1fd890ac1e0 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0frt-9147300000-0df80e53ff7eae16ec1d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000i-0000900000-9023eec8b578fa8dcd3c | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-000i-0000900000-be644c278ee03382b0e0 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00di-3003900000-00a5c7f7178a091e67af | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000i-0000900000-6abf4200d93b5ca719d0 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-000i-0000900000-211f179674261b0fdd6b | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0079-0001900000-608b1ef50efb487ffb7d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03di-1918600000-b1c4f0a0bf89ec7a8d17 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-053s-9211100000-826cb673f380da85d9cf | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0bu4-9712000000-cc344701627ecac3244a | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0034190 |
|---|
| FooDB ID | FDB012484 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | Not Available |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 131751533 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|