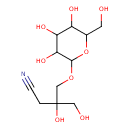
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-08i0-6490000000-ff8c3f61f47dddbeb71f | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (5 TMS) - 70eV, Positive | splash10-000i-1231039000-3b7930a14880a3c5626d | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-01tc-1390000000-94aada39a91a85a8f8b3 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03dl-3930000000-e2096e58b4742d027fbe | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03dm-9800000000-d156848eda02249208c3 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-01ox-2790000000-b645c838d5fdf60e8b63 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-03kc-6950000000-92b0855e4082c8aaaeef | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9300000000-0e5d109a1519441a9b6a | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-01ox-1790000000-53189ffc47cb3152e8fd | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-055f-3940000000-a7e189fea8422907a542 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a5c-9200000000-5de95052d0a3c37612b9 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-01ox-0690000000-48ae3b7209d8f3210859 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03ka-9600000000-e57c2df815db5808ba82 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0006-9200000000-a798793dcbcf46a072a8 | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |