| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 00:17:06 UTC |
|---|
| Update Date | 2016-11-09 01:18:38 UTC |
|---|
| Accession Number | CHEM028196 |
|---|
| Identification |
|---|
| Common Name | Brassinolide |
|---|
| Class | Small Molecule |
|---|
| Description | |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
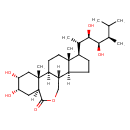
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 24-Epibrassinolide | ChEMBL, HMDB | | 2,3,22,23-Tetrahydroxy-24-methyl-b-homo-7-oxacholestan-6-one | HMDB | | 2alpha,3alpha,22alpha,23alpha-Tetrahydroxy-24alpha-methyl-b-homo-7-oxa-5alpha-cholestan-6-one | MeSH, HMDB | | Brassinolide, (2alpha,3alpha,5alpha,22S,23S)-isomer | MeSH, HMDB | | Brassinolide, (2alpha,3alpha,5alpha.22R,23R)-isomer | MeSH, HMDB | | Brassinolide | MeSH |
|
|---|
| Chemical Formula | C28H48O6 |
|---|
| Average Molecular Mass | 480.677 g/mol |
|---|
| Monoisotopic Mass | 480.345 g/mol |
|---|
| CAS Registry Number | 72962-43-7 |
|---|
| IUPAC Name | (1S,2R,4R,5S,7S,11S,12S,15R,16S)-15-[(2S,3R,4R,5R)-3,4-dihydroxy-5,6-dimethylheptan-2-yl]-4,5-dihydroxy-2,16-dimethyl-9-oxatetracyclo[9.7.0.0²,⁷.0¹²,¹⁶]octadecan-8-one |
|---|
| Traditional Name | 24-epi-brassinolide |
|---|
| SMILES | [H][C@@]12CC[C@H]([C@H](C)[C@@H](O)[C@H](O)[C@H](C)C(C)C)[C@@]1(C)CC[C@@]1([H])[C@@]2([H])COC(=O)[C@@]2([H])C[C@H](O)[C@H](O)C[C@]12C |
|---|
| InChI Identifier | InChI=1S/C28H48O6/c1-14(2)15(3)24(31)25(32)16(4)18-7-8-19-17-13-34-26(33)21-11-22(29)23(30)12-28(21,6)20(17)9-10-27(18,19)5/h14-25,29-32H,7-13H2,1-6H3/t15-,16+,17+,18-,19+,20+,21-,22+,23-,24-,25-,27-,28-/m1/s1 |
|---|
| InChI Key | IXVMHGVQKLDRKH-QHBHMFGVSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as brassinolides and derivatives. These are cholestane based steroid lactones containing benzo[c]indeno[5,4-e]oxepin-3-one. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Steroids and steroid derivatives |
|---|
| Sub Class | Steroid lactones |
|---|
| Direct Parent | Brassinolides and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Brassinolide-skeleton
- Caprolactone
- Oxepane
- Cyclic alcohol
- Carboxylic acid ester
- Lactone
- Secondary alcohol
- Carboxylic acid derivative
- Oxacycle
- Organoheterocyclic compound
- Monocarboxylic acid or derivatives
- Hydrocarbon derivative
- Alcohol
- Carbonyl group
- Organic oxygen compound
- Organic oxide
- Organooxygen compound
- Aliphatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aliphatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0ir3-4315900000-c0cdd9a5e00ed559b037 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (3 TMS) - 70eV, Positive | splash10-001i-3111209000-19e3d547be0a70e9894d | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-01q9-0001900000-f751356e978d7611841d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0irs-7209600000-b9396588d263ce2fe7e3 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0f89-9506200000-0d91d1fed47d6d5f72cf | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-0000900000-52b84cac64a5ac86adbf | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0200-3204900000-88c87f58b6d8d294c790 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00fs-9405300000-065785cd0b58a9979545 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-0000900000-b27078fda0520f837b35 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004j-2305900000-f2b2e8bcfa7706e2375d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-056r-3005900000-b530898f59fc290c0951 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-001j-0009600000-c36102c0fe9ca31dc76b | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-008a-4309100000-2ca728d9dc9e0fbfb809 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4l-9415000000-36c9809f108ce77f9364 | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0034081 |
|---|
| FooDB ID | FDB012340 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | C00000180 |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Brassinolide |
|---|
| Chemspider ID | 391354 |
|---|
| ChEBI ID | 27722 |
|---|
| PubChem Compound ID | 443055 |
|---|
| Kegg Compound ID | C11049 |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|