| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-26 00:02:51 UTC |
|---|
| Update Date | 2016-11-09 01:18:33 UTC |
|---|
| Accession Number | CHEM027877 |
|---|
| Identification |
|---|
| Common Name | Desglucocoroloside |
|---|
| Class | Small Molecule |
|---|
| Description | Desglucocoroloside is found in green vegetables. Desglucocoroloside is from Corchorus olitorius (Jew's mallow). |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
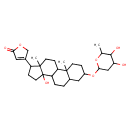
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Deglucocoroloside | HMDB | | Digitoxigenin boivinoside | HMDB |
|
|---|
| Chemical Formula | C29H44O7 |
|---|
| Average Molecular Mass | 504.656 g/mol |
|---|
| Monoisotopic Mass | 504.309 g/mol |
|---|
| CAS Registry Number | 57361-72-5 |
|---|
| IUPAC Name | 4-{5-[(4,5-dihydroxy-6-methyloxan-2-yl)oxy]-11-hydroxy-2,15-dimethyltetracyclo[8.7.0.0²,⁷.0¹¹,¹⁵]heptadecan-14-yl}-2,5-dihydrofuran-2-one |
|---|
| Traditional Name | 4-{5-[(4,5-dihydroxy-6-methyloxan-2-yl)oxy]-11-hydroxy-2,15-dimethyltetracyclo[8.7.0.0²,⁷.0¹¹,¹⁵]heptadecan-14-yl}-5H-furan-2-one |
|---|
| SMILES | CC1OC(CC(O)C1O)OC1CCC2(C)C(CCC3C2CCC2(C)C(CCC32O)C2=CC(=O)OC2)C1 |
|---|
| InChI Identifier | InChI=1S/C29H44O7/c1-16-26(32)23(30)14-25(35-16)36-19-6-9-27(2)18(13-19)4-5-22-21(27)7-10-28(3)20(8-11-29(22,28)33)17-12-24(31)34-15-17/h12,16,18-23,25-26,30,32-33H,4-11,13-15H2,1-3H3 |
|---|
| InChI Key | NQOMDNMTNVQXRR-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as cardenolide glycosides and derivatives. Cardenolide glycosides and derivatives are compounds containing a carbohydrate glycosidically bound to the cardenolide moiety. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Steroids and steroid derivatives |
|---|
| Sub Class | Steroid lactones |
|---|
| Direct Parent | Cardenolide glycosides and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Cardanolide-glycoside
- Steroidal glycoside
- 14-hydroxysteroid
- Hydroxysteroid
- Hexose monosaccharide
- Glycosyl compound
- O-glycosyl compound
- 2-furanone
- Monosaccharide
- Oxane
- Cyclic alcohol
- Dihydrofuran
- Alpha,beta-unsaturated carboxylic ester
- Enoate ester
- Tertiary alcohol
- Lactone
- Carboxylic acid ester
- Secondary alcohol
- Organoheterocyclic compound
- Oxacycle
- Acetal
- Carboxylic acid derivative
- Monocarboxylic acid or derivatives
- Hydrocarbon derivative
- Alcohol
- Organic oxide
- Organic oxygen compound
- Carbonyl group
- Organooxygen compound
- Aliphatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aliphatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-004r-2326900000-474ce68d2e2667946e42 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-0f89-4322219000-175918452b9751d817a0 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0a70-0006920000-d8bc2ba3b2263214225b | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a59-0209100000-3fd536fc4d27315db3ad | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-001l-1935000000-3a126c6e98278976e0c4 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0zfr-0107790000-c25493a7ce294cf4ae77 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0aba-1109200000-089c3fad5e1dd6cb2376 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-05fr-2009000000-ec2f4eadaa37b5c0c657 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0uk9-0008690000-1eaf0c345e01dae5e8bb | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0fk9-4209340000-4a654cedf2a8fc041942 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0007-9107100000-4f790fd3734debe89aaa | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0a4r-0029870000-27a7470fe23fa1bb6a99 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0avr-0387930000-676e2bd7f70fe43d7309 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-05r0-6944000000-935852a59ee1e9945acd | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0033709 |
|---|
| FooDB ID | FDB011825 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 10444697 |
|---|
| ChEBI ID | 173218 |
|---|
| PubChem Compound ID | 15559187 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|