| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-25 23:55:20 UTC |
|---|
| Update Date | 2016-11-09 01:18:30 UTC |
|---|
| Accession Number | CHEM027702 |
|---|
| Identification |
|---|
| Common Name | (9Z,11R,12S,13S,15Z)-12,13-Epoxy-11-hydroxy-9,15-octadecadienoic acid |
|---|
| Class | Small Molecule |
|---|
| Description | (9Z,11S,12S,13S,12Z)-12,13-Epoxy-11-hydroxy-9,15-octadecadienoic acid is isolated from leaves of rice plants with rice blast disease (Pyricularia oryzae). |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
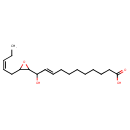
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (9Z,11R,12S,13S,15Z)-12,13-Epoxy-11-hydroxy-9,15-octadecadienoate | Generator | | (9E)-11-Hydroxy-11-{3-[(2Z)-pent-2-en-1-yl]oxiran-2-yl}undec-9-enoate | HMDB |
|
|---|
| Chemical Formula | C18H30O4 |
|---|
| Average Molecular Mass | 310.428 g/mol |
|---|
| Monoisotopic Mass | 310.214 g/mol |
|---|
| CAS Registry Number | 106034-49-5 |
|---|
| IUPAC Name | (9E)-11-hydroxy-11-{3-[(2Z)-pent-2-en-1-yl]oxiran-2-yl}undec-9-enoic acid |
|---|
| Traditional Name | (9E)-11-hydroxy-11-{3-[(2Z)-pent-2-en-1-yl]oxiran-2-yl}undec-9-enoic acid |
|---|
| SMILES | CC\C=C/CC1OC1C(O)\C=C\CCCCCCCC(O)=O |
|---|
| InChI Identifier | InChI=1S/C18H30O4/c1-2-3-9-13-16-18(22-16)15(19)12-10-7-5-4-6-8-11-14-17(20)21/h3,9-10,12,15-16,18-19H,2,4-8,11,13-14H2,1H3,(H,20,21)/b9-3-,12-10+ |
|---|
| InChI Key | XHMGEKKHSKIGMI-JCMSFEFZSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as long-chain fatty acids. These are fatty acids with an aliphatic tail that contains between 13 and 21 carbon atoms. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Fatty Acyls |
|---|
| Sub Class | Fatty acids and conjugates |
|---|
| Direct Parent | Long-chain fatty acids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Long-chain fatty acid
- Epoxy fatty acid
- Heterocyclic fatty acid
- Hydroxy fatty acid
- Unsaturated fatty acid
- Secondary alcohol
- Monocarboxylic acid or derivatives
- Oxacycle
- Ether
- Oxirane
- Dialkyl ether
- Carboxylic acid
- Carboxylic acid derivative
- Organoheterocyclic compound
- Alcohol
- Organooxygen compound
- Hydrocarbon derivative
- Organic oxide
- Organic oxygen compound
- Carbonyl group
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-00l2-5920000000-d401b41186cd6db29f36 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-00mx-9346200000-2c61761ce5c0b711fe4d | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03dl-5397000000-af7c490b2ea5ad9b4ffd | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-001i-9610000000-230bb2911295e417fac6 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-100u-9200000000-7a54a6d9d362dac77a99 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0529000000-e0a11e6dc2fc8f87522c | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0cdi-1932000000-edf278cc291da170a466 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9200000000-90a46a1a875e4be2fad6 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03fu-2693000000-e2e1b03e12a16f4f639f | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-06ur-9871000000-2c9564957510cd87ced7 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-05mo-9200000000-9c522ace71775345c5ad | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0029000000-76036cb86db5f0c5083c | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4l-2595000000-44b4f3d7acdcb536ef4e | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9430000000-f5c404823c63d99c8d49 | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0033505 |
|---|
| FooDB ID | FDB011557 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 35013615 |
|---|
| ChEBI ID | 138784 |
|---|
| PubChem Compound ID | 131751443 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|