| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-25 23:49:23 UTC |
|---|
| Update Date | 2016-11-09 01:18:29 UTC |
|---|
| Accession Number | CHEM027569 |
|---|
| Identification |
|---|
| Common Name | 2,3-Epoxysesamone |
|---|
| Class | Small Molecule |
|---|
| Description | 2,3-Epoxysesamone is found in fats and oils. 2,3-Epoxysesamone is a constituent of Sesamum indicum (sesame). |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
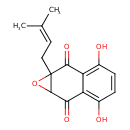
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 2,3-Epoxy-2-prenylnaphthazarin | HMDB | | 3,6-Dihydroxy-1a-(3-methyl-2-butenyl)naphth[2,3-b]-2,7(1ah,7ah)-dione | HMDB | | 2,3-Epoxy-2,3-dihydro-5,8-dihydroxy-2-(3-methyl-2-butenyl)-1,4-naphthoquinone | MeSH, HMDB | | 2,3-Epoxysesamone | MeSH |
|
|---|
| Chemical Formula | C15H14O5 |
|---|
| Average Molecular Mass | 274.269 g/mol |
|---|
| Monoisotopic Mass | 274.084 g/mol |
|---|
| CAS Registry Number | 111261-12-2 |
|---|
| IUPAC Name | 3,6-dihydroxy-1a-(3-methylbut-2-en-1-yl)-1aH,2H,7H,7aH-naphtho[2,3-b]oxirene-2,7-dione |
|---|
| Traditional Name | 3,6-dihydroxy-1a-(3-methylbut-2-en-1-yl)-7aH-naphtho[2,3-b]oxirene-2,7-dione |
|---|
| SMILES | CC(C)=CCC12OC1C(=O)C1=C(O)C=CC(O)=C1C2=O |
|---|
| InChI Identifier | InChI=1S/C15H14O5/c1-7(2)5-6-15-13(19)11-9(17)4-3-8(16)10(11)12(18)14(15)20-15/h3-5,14,16-17H,6H2,1-2H3 |
|---|
| InChI Key | TWESJUCJKWFWQV-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as vitamin k compounds. These are quinone lipids containing a methylated naphthoquinone ring structure, and vary in the aliphatic side chain attached at the 3-position. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Quinone and hydroquinone lipids |
|---|
| Direct Parent | Vitamin K compounds |
|---|
| Alternative Parents | |
|---|
| Substituents | - Naphthoquinone
- Naphthalene
- Tetralin
- Quinone
- Aryl ketone
- Aryl alkyl ketone
- 1-hydroxy-2-unsubstituted benzenoid
- Benzenoid
- Vinylogous acid
- Ketone
- Organoheterocyclic compound
- Oxacycle
- Ether
- Oxirane
- Dialkyl ether
- Hydrocarbon derivative
- Organooxygen compound
- Organic oxide
- Organic oxygen compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0aou-9660000000-cc32369a36830feaf47e | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-0690-9735200000-14011ee3096da8f0ee02 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-004i-0090000000-cb7e1c6f1df7d049f188 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-016r-3190000000-8a2a4b4f692d23ee9e7e | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0909-7970000000-38d2113c12e3bac30619 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00di-0090000000-e17d6e478a0a75ffb681 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-05fr-1190000000-567e77ce1e40f87655ec | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-08fr-4910000000-dc98bfcb80f25af0f2c3 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-004i-0090000000-0ff4f807d4047e6d45c3 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-004i-0190000000-c5ccc35be7fb86c6ca5a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-016u-4690000000-0a397f113b10452fe25e | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00di-0090000000-cdeef1cb9497dc9eee76 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00di-0890000000-dd7868e3c07581671081 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-05fv-5390000000-3c419a891c42c4712eb7 | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0033333 |
|---|
| FooDB ID | FDB011360 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | C00056462 |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 320321 |
|---|
| ChEBI ID | 174606 |
|---|
| PubChem Compound ID | 360837 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|