| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-25 23:25:32 UTC |
|---|
| Update Date | 2016-11-09 01:18:22 UTC |
|---|
| Accession Number | CHEM026986 |
|---|
| Identification |
|---|
| Common Name | Chatenaytrienin 2 |
|---|
| Class | Small Molecule |
|---|
| Description | Chatenaytrienin 2 is found in fruits. Chatenaytrienin 2 is a constituent of Annona muricata (soursop) |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
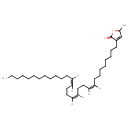
| Chemical Structure | |
|---|
| Synonyms | Not Available |
|---|
| Chemical Formula | C35H60O2 |
|---|
| Average Molecular Mass | 512.850 g/mol |
|---|
| Monoisotopic Mass | 512.459 g/mol |
|---|
| CAS Registry Number | 206131-74-0 |
|---|
| IUPAC Name | 5-methyl-3-[(9Z,13Z,17Z)-triaconta-9,13,17-trien-1-yl]-2,5-dihydrofuran-2-one |
|---|
| Traditional Name | 5-methyl-3-[(9Z,13Z,17Z)-triaconta-9,13,17-trien-1-yl]-5H-furan-2-one |
|---|
| SMILES | CCCCCCCCCCCC\C=C/CC\C=C/CC\C=C/CCCCCCCCC1=CC(C)OC1=O |
|---|
| InChI Identifier | InChI=1S/C35H60O2/c1-3-4-5-6-7-8-9-10-11-12-13-14-15-16-17-18-19-20-21-22-23-24-25-26-27-28-29-30-31-34-32-33(2)37-35(34)36/h14-15,18-19,22-23,32-33H,3-13,16-17,20-21,24-31H2,1-2H3/b15-14-,19-18-,23-22- |
|---|
| InChI Key | GTXRLWWQVRDMLK-VCHJKVOASA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as annonaceous acetogenins. These are waxy derivatives of fatty acids (usually C32 or C34), containing a terminal carboxylic acid combined with a 2-propanol unit at the C-2 position to form a methyl- substituted alpha,beta-unsaturated-gamma-lactone. One of their interesting structural features is a single, adjacent, or nonadjacent tetrahydrofuran (THF) or tetrahydropyran (THP) system with one or two flanking hydroxyl group(s) at the center of a long hydrocarbon chain. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Fatty Acyls |
|---|
| Sub Class | Fatty alcohols |
|---|
| Direct Parent | Annonaceous acetogenins |
|---|
| Alternative Parents | |
|---|
| Substituents | - Annonaceae acetogenin skeleton
- 2-furanone
- Dihydrofuran
- Alpha,beta-unsaturated carboxylic ester
- Enoate ester
- Carboxylic acid ester
- Lactone
- Carboxylic acid derivative
- Oxacycle
- Organoheterocyclic compound
- Monocarboxylic acid or derivatives
- Hydrocarbon derivative
- Organic oxygen compound
- Organic oxide
- Carbonyl group
- Organooxygen compound
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0pba-4976200000-4800ca4a7958c36fc4e2 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03di-0010590000-185e1890afb61338dd15 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0gi0-0240900000-6c2ddd436c221f672e6b | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00to-1101900000-23f236901352ff8fa3d7 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03di-1000390000-2be8c2de3a86190f1fa4 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-03xr-2000970000-1c9bbb4721eb5e14c429 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-014r-3000900000-a191138b2f6662132f5d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03di-0000090000-a549e76c6d174827f8ca | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-03di-2000490000-15c841f3f288edf9e804 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-01b9-4400900000-63d0d3e54150f9dcc8e6 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03di-0111390000-861c4180bb08c4d2784f | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03xs-2000930000-05b98837d72f7d488fc9 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00lu-9615600000-24a0ec07b3cd65bd42ba | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0032647 |
|---|
| FooDB ID | FDB010596 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | C00048960 |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 35013460 |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 131751272 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|