| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-25 23:15:58 UTC |
|---|
| Update Date | 2016-11-09 01:18:20 UTC |
|---|
| Accession Number | CHEM026834 |
|---|
| Identification |
|---|
| Common Name | Gibberellin A112 |
|---|
| Class | Small Molecule |
|---|
| Description | Constituent of Matthiola incana and Raphanus sativus (radish). Gibberellin A112 is found in strawberry, root vegetables, and radish. |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
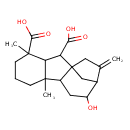
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| GA112 | HMDB | | 11-Hydroxy-4,8-dimethyl-13-methylidenetetracyclo[10.2.1.0¹,⁹.0³,⁸]pentadecane-2,4-dicarboxylate | Generator |
|
|---|
| Chemical Formula | C20H28O5 |
|---|
| Average Molecular Mass | 348.433 g/mol |
|---|
| Monoisotopic Mass | 348.194 g/mol |
|---|
| CAS Registry Number | 90806-15-8 |
|---|
| IUPAC Name | 11-hydroxy-4,8-dimethyl-13-methylidenetetracyclo[10.2.1.0¹,⁹.0³,⁸]pentadecane-2,4-dicarboxylic acid |
|---|
| Traditional Name | 11-hydroxy-4,8-dimethyl-13-methylidenetetracyclo[10.2.1.0¹,⁹.0³,⁸]pentadecane-2,4-dicarboxylic acid |
|---|
| SMILES | CC12CCCC(C)(C1C(C(O)=O)C13CC(C(O)CC21)C(=C)C3)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C20H28O5/c1-10-8-20-9-11(10)12(21)7-13(20)18(2)5-4-6-19(3,17(24)25)15(18)14(20)16(22)23/h11-15,21H,1,4-9H2,2-3H3,(H,22,23)(H,24,25) |
|---|
| InChI Key | RVQCZHZIMZMGAD-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as c20-gibberellin 6-carboxylic acids. These are c20-gibberellins with a carboxyl group at the 6-position. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Diterpenoids |
|---|
| Direct Parent | C20-gibberellin 6-carboxylic acids |
|---|
| Alternative Parents | Not Available |
|---|
| Substituents | Not Available |
|---|
| Molecular Framework | Aliphatic homopolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0ue9-2229000000-dca6d0152873846d3045 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (3 TMS) - 70eV, Positive | splash10-002b-3031970000-e0bcf6545dac18e115d6 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-001j-0029000000-47e4ee9b71d887b496ff | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0gwr-0069000000-3c041d02d8018484873f | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-052r-2592000000-b3d0b69aa8e4a42362cd | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0f6t-0029000000-00d79a251d0632bf800c | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0zg1-0079000000-d4002d55a8d1339a51c5 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4r-1094000000-108c195a785191117608 | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0032011 |
|---|
| FooDB ID | FDB008709 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | C00001210 |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | Not Available |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 131751245 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|