| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-25 23:14:48 UTC |
|---|
| Update Date | 2016-11-09 01:18:20 UTC |
|---|
| Accession Number | CHEM026804 |
|---|
| Identification |
|---|
| Common Name | Methyl (9Z)-8'-oxo-6,8'-diapo-6-carotenoate |
|---|
| Class | Small Molecule |
|---|
| Description | Methyl (9Z)-8'-oxo-6,8'-diapo-6-carotenoate is a constituent of Bixa orellana (annatto) |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
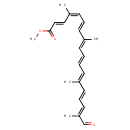
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Methyl (9Z)-8'-oxo-6,8'-diapo-6-carotenoic acid | Generator | | apo-8'-Bixinal | HMDB | | Methyl (4Z,6E,8E,10E,12E,14E,16E)-4,8,13,17-tetramethyl-18-oxooctadeca-2,4,6,8,10,12,14,16-octaenoic acid | Generator |
|
|---|
| Chemical Formula | C23H28O3 |
|---|
| Average Molecular Mass | 352.467 g/mol |
|---|
| Monoisotopic Mass | 352.204 g/mol |
|---|
| CAS Registry Number | 101034-51-9 |
|---|
| IUPAC Name | methyl (2E,4Z,6E,8E,10E,12E,14E,16E)-4,8,13,17-tetramethyl-18-oxooctadeca-2,4,6,8,10,12,14,16-octaenoate |
|---|
| Traditional Name | methyl (2E,4Z,6E,8E,10E,12E,14E,16E)-4,8,13,17-tetramethyl-18-oxooctadeca-2,4,6,8,10,12,14,16-octaenoate |
|---|
| SMILES | COC(=O)\C=C\C(\C)=C/C=C/C(/C)=C/C=C/C=C(\C)/C=C/C=C(\C)C=O |
|---|
| InChI Identifier | InChI=1S/C23H28O3/c1-19(12-8-14-21(3)16-17-23(25)26-5)10-6-7-11-20(2)13-9-15-22(4)18-24/h6-18H,1-5H3/b7-6+,12-8+,13-9+,17-16+,19-10+,20-11+,21-14-,22-15+ |
|---|
| InChI Key | JMFNGSBVJBPGQR-IWRWKHRWSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as acyclic diterpenoids. These are diterpenoids (compounds made of four consecutive isoprene units) that do not contain a cycle. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Diterpenoids |
|---|
| Direct Parent | Acyclic diterpenoids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Acyclic diterpenoid
- Octadecanoid
- Fatty aldehyde
- Fatty acid ester
- Fatty acyl
- Alpha,beta-unsaturated aldehyde
- Enal
- Alpha,beta-unsaturated carboxylic ester
- Methyl ester
- Enoate ester
- Carboxylic acid ester
- Carboxylic acid derivative
- Monocarboxylic acid or derivatives
- Hydrocarbon derivative
- Organic oxide
- Aldehyde
- Organooxygen compound
- Organic oxygen compound
- Carbonyl group
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0079-0359000000-17b8b9040bba342d007e | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0fk9-1139000000-d92b0515cfa644005f5e | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-06gu-1691000000-aa7da177bcb203ca9794 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-05nb-5950000000-913a9a523da4d469b729 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0udi-0009000000-c1117c17716f0c5458bf | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0uxr-1019000000-de53a03d209a45bcd5b2 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-9145000000-2e6da48c8da6852fb798 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0udi-0059000000-776aeb67ea085156c3ed | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0lk9-4098000000-e37bc2636a45d41b51b9 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-000i-0290000000-a0d48b745e82375a01ed | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-01c3-0092000000-3e4fbfcddc71c21986ea | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-014u-2291000000-4b93860da2dfd9065c8f | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4i-3960000000-c84aeb1f836de338287c | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0031979 |
|---|
| FooDB ID | FDB008672 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | C00023149 |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 9221571 |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 11046403 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|