| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-25 23:12:17 UTC |
|---|
| Update Date | 2016-11-09 01:18:19 UTC |
|---|
| Accession Number | CHEM026734 |
|---|
| Identification |
|---|
| Common Name | Porric acid B |
|---|
| Class | Small Molecule |
|---|
| Description | Constituent of Annona muricata (soursop). Muridienin 4 is found in fruits. |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
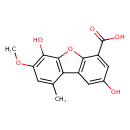
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 3-(15,19-Dotriacontadienyl)-5-methyl-2(5H)-furanone | HMDB | | 2,6-Dihydroxy-7-methoxy-9-methyl-4-dibenzofurancarboxylic acid | HMDB | | 4,10-Dihydroxy-11-methoxy-13-methyl-8-oxatricyclo[7.4.0.0²,⁷]trideca-1(9),2(7),3,5,10,12-hexaene-6-carboxylate | Generator | | Porrate b | Generator |
|
|---|
| Chemical Formula | C15H12O6 |
|---|
| Average Molecular Mass | 288.252 g/mol |
|---|
| Monoisotopic Mass | 288.063 g/mol |
|---|
| CAS Registry Number | 207285-02-7 |
|---|
| IUPAC Name | 4,10-dihydroxy-11-methoxy-13-methyl-8-oxatricyclo[7.4.0.0²,⁷]trideca-1(9),2(7),3,5,10,12-hexaene-6-carboxylic acid |
|---|
| Traditional Name | 4,10-dihydroxy-11-methoxy-13-methyl-8-oxatricyclo[7.4.0.0²,⁷]trideca-1(9),2(7),3,5,10,12-hexaene-6-carboxylic acid |
|---|
| SMILES | COC1=C(O)C2=C(C3=C(O2)C(=CC(O)=C3)C(O)=O)C(C)=C1 |
|---|
| InChI Identifier | InChI=1S/C15H12O6/c1-6-3-10(20-2)12(17)14-11(6)8-4-7(16)5-9(15(18)19)13(8)21-14/h3-5,16-17H,1-2H3,(H,18,19) |
|---|
| InChI Key | YRDAFVVZXCAVIU-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as annonaceous acetogenins. These are waxy derivatives of fatty acids (usually C32 or C34), containing a terminal carboxylic acid combined with a 2-propanol unit at the C-2 position to form a methyl- substituted alpha,beta-unsaturated-gamma-lactone. One of their interesting structural features is a single, adjacent, or nonadjacent tetrahydrofuran (THF) or tetrahydropyran (THP) system with one or two flanking hydroxyl group(s) at the center of a long hydrocarbon chain. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Fatty Acyls |
|---|
| Sub Class | Fatty alcohols |
|---|
| Direct Parent | Annonaceous acetogenins |
|---|
| Alternative Parents | |
|---|
| Substituents | - Annonaceae acetogenin skeleton
- 2-furanone
- Dihydrofuran
- Alpha,beta-unsaturated carboxylic ester
- Enoate ester
- Carboxylic acid ester
- Lactone
- Carboxylic acid derivative
- Oxacycle
- Organoheterocyclic compound
- Monocarboxylic acid or derivatives
- Hydrocarbon derivative
- Organic oxygen compound
- Organic oxide
- Carbonyl group
- Organooxygen compound
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-00dl-0190000000-70242ef807b2aa58930b | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (3 TMS) - 70eV, Positive | splash10-001c-1201900000-7e0b6d5b1491a3570cb4 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-0090000000-838852ec44bfe85d02b7 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0076-0090000000-d70ed097eaf6016a9af6 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-006x-1190000000-6ae3aa7542f2a4503db4 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000l-0090000000-344597db8aecae27ba43 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-002f-0090000000-76718f4ccdf074aabf7c | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-0290000000-b7668b23dc045c5501a2 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0079-0090000000-6081a931933f97746b48 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00dr-0090000000-4a9fbcb449552fa6785c | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-02vl-0290000000-d6c09985b1d3449cae67 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000i-0090000000-74127368fd472a94be09 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-03g3-0090000000-b9b949e8f1d38cd652d4 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-01t9-0090000000-d5f9798732476e330d08 | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0031906 |
|---|
| FooDB ID | FDB008593 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | Not Available |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 131751208 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|